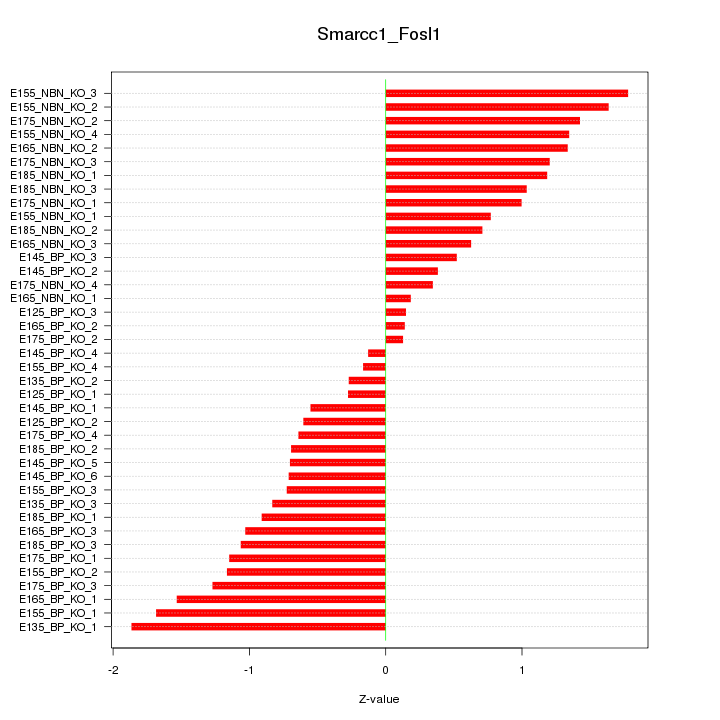
Motif ID: Smarcc1_Fosl1
Z-value: 0.981
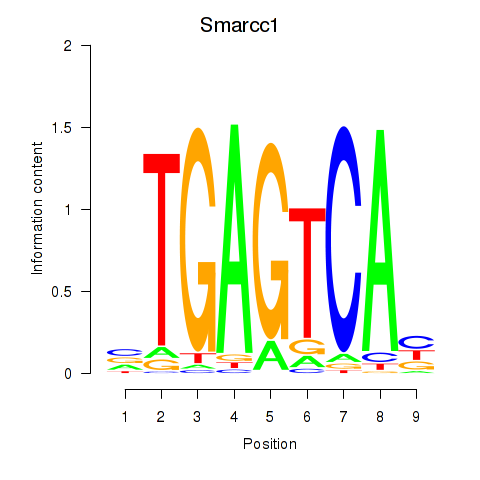
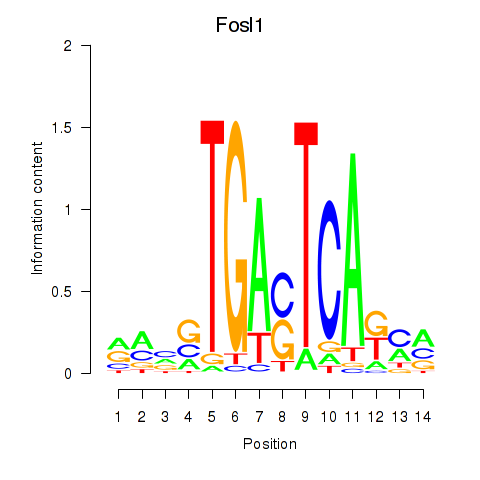
Transcription factors associated with Smarcc1_Fosl1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Fosl1 | ENSMUSG00000024912.5 | Fosl1 |
| Smarcc1 | ENSMUSG00000032481.10 | Smarcc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosl1 | mm10_v2_chr19_+_5447692_5447711 | 0.70 | 4.6e-07 | Click! |
| Smarcc1 | mm10_v2_chr9_+_110132015_110132102 | -0.67 | 2.0e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.5 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.1 | 3.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.0 | 3.0 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.0 | 4.8 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.9 | 8.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.9 | 4.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.9 | 2.6 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.9 | 6.0 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.8 | 2.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.8 | 3.9 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.8 | 3.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.7 | 2.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.7 | 2.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.7 | 2.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 0.7 | 3.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 2.0 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.7 | 2.6 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.6 | 3.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.6 | 1.9 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.6 | 2.4 | GO:0035793 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.6 | 1.7 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.5 | 2.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 1.6 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.5 | 3.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.5 | 2.6 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.5 | 0.5 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.5 | 1.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.5 | 1.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 1.4 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.5 | 2.8 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 | 3.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.5 | 2.3 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.4 | 1.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 2.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 0.9 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.4 | 1.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.4 | 0.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.4 | 2.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.4 | 0.4 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.4 | 1.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 2.5 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.4 | 1.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 4.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 1.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 1.2 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.4 | 1.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 2.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 0.8 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.4 | 1.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 5.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 1.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 0.7 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.4 | 1.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.4 | 1.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 1.4 | GO:1900122 | transformation of host cell by virus(GO:0019087) positive regulation of receptor binding(GO:1900122) |
| 0.4 | 1.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.4 | 0.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 1.0 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.3 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.3 | 1.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 1.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 2.0 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.3 | 2.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.3 | 1.0 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.3 | 1.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 1.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 0.9 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.3 | 2.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.3 | 0.9 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 1.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 1.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 0.8 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.3 | 1.6 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 3.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 2.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.3 | 1.3 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.3 | 0.5 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.3 | 4.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 1.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 1.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 1.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 12.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 1.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 1.5 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 0.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 1.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 2.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.2 | 1.9 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 0.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 0.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 1.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.2 | 1.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 2.4 | GO:0032328 | alanine transport(GO:0032328) |
| 0.2 | 0.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 0.2 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 | 1.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 1.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.2 | 1.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 2.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.8 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 7.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 0.8 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.2 | 2.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 1.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.6 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 1.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 1.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.2 | 0.9 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.2 | 0.6 | GO:0061744 | motor behavior(GO:0061744) |
| 0.2 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 1.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 0.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 0.9 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 0.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 0.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.7 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.2 | 1.3 | GO:0061718 | NADH regeneration(GO:0006735) regulation of anion channel activity(GO:0010359) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.1 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 | 1.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 1.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 0.7 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.2 | 0.3 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 1.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.7 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 1.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.7 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 1.6 | GO:0019243 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.3 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.2 | 0.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 0.3 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.2 | 0.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.5 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.8 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 3.6 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 0.4 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.4 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.9 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 5.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.4 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 1.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.4 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.1 | 1.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.6 | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell(GO:1904994) |
| 0.1 | 0.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 1.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 1.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 6.1 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 0.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 1.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.1 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 1.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.9 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.8 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.9 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.3 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.4 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 0.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.5 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.7 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.3 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 1.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 2.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.5 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 0.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.7 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.6 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.6 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.3 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.1 | 0.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.6 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.5 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.7 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.9 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.3 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.7 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.6 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 3.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.4 | GO:0009227 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.7 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.7 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 2.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 1.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.3 | GO:1903764 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 5.2 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.6 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.2 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 1.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.4 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.2 | GO:1904046 | seryl-tRNA aminoacylation(GO:0006434) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.1 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 2.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.6 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.2 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.3 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.6 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 3.1 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.1 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.2 | GO:0018202 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 2.1 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 0.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.7 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.4 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 2.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.5 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:1902083 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.3 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) Golgi reassembly(GO:0090168) |
| 0.0 | 0.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.1 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.7 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.8 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.7 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 1.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.2 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.9 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.1 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.0 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.0 | 0.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.4 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.5 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.2 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.4 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 1.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 | 1.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:0031034 | myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.4 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.2 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.2 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 0.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.2 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.0 | 3.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 2.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.6 | 2.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 1.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.6 | 1.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.6 | 6.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.6 | 2.3 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.5 | 3.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 2.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 1.9 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 1.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 2.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.3 | 6.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 1.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 0.9 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.3 | 3.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 11.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 0.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.2 | 0.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 2.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 0.7 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) |
| 0.2 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 4.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 2.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 3.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 3.0 | GO:0032421 | stereocilium(GO:0032420) stereocilium bundle(GO:0032421) |
| 0.2 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 1.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 0.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 0.7 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 4.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 4.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 3.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 4.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 8.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 4.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 9.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 7.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 22.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.2 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 2.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 5.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.2 | GO:0000802 | transverse filament(GO:0000802) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.2 | GO:0044301 | climbing fiber(GO:0044301) parallel fiber(GO:1990032) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 5.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 1.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 3.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 1.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 2.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.7 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.5 | 6.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.4 | 4.2 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 1.1 | 3.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 2.6 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.8 | 6.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 2.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 2.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.6 | 2.5 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.6 | 2.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 6.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 1.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 2.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 3.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 1.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 3.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 1.2 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.4 | 1.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.4 | 3.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.4 | 1.9 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.4 | 2.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 1.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 1.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 1.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 1.4 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.3 | 5.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 8.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 1.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.0 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.3 | 1.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 2.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 1.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 0.9 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.3 | 2.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 1.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 0.8 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.3 | 1.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 0.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 1.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 4.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 1.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 1.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.7 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 1.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 1.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 7.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 0.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 0.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 0.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 1.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 1.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 0.7 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 2.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 0.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 7.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 4.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.6 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 2.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 3.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.4 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 3.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 4.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 4.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 2.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0038132 | receptor signaling protein tyrosine kinase activity(GO:0004716) neuregulin binding(GO:0038132) |
| 0.1 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.1 | 2.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.6 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 2.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.5 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 3.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 3.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 2.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 8.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 3.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 3.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.6 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 6.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 2.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.0 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 3.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 6.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 1.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.5 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.4 | 0.4 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 7.6 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.2 | 0.9 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.2 | 3.9 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 10.0 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 2.9 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 6.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.1 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.7 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.0 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.0 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.1 | 7.3 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.9 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.8 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.5 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.1 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 11.7 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.9 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.4 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.1 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.4 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.0 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 0.8 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.9 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 4.1 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.3 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.5 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.8 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID_REELIN_PATHWAY | Reelin signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 4.0 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 7.3 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 2.5 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 0.8 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 3.8 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 3.7 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.0 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 8.6 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 7.1 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 0.4 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 2.8 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 4.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 5.7 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 0.8 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.7 | REACTOME_SHC_MEDIATED_SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.8 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.6 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.1 | REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 5.2 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.8 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.0 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.2 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.3 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.8 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.1 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.8 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.6 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.8 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.3 | REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 0.7 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.0 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 7.8 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.1 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.5 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 5.5 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.9 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.4 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.5 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.8 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.1 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.7 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.6 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.7 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.8 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.2 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.4 | REACTOME_REGULATION_OF_APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.0 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.0 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 4.5 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.5 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.4 | REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 2.9 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.6 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.2 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.1 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.1 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.0 | REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.3 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.9 | REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.5 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.6 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.7 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME_LIPOPROTEIN_METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.3 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |