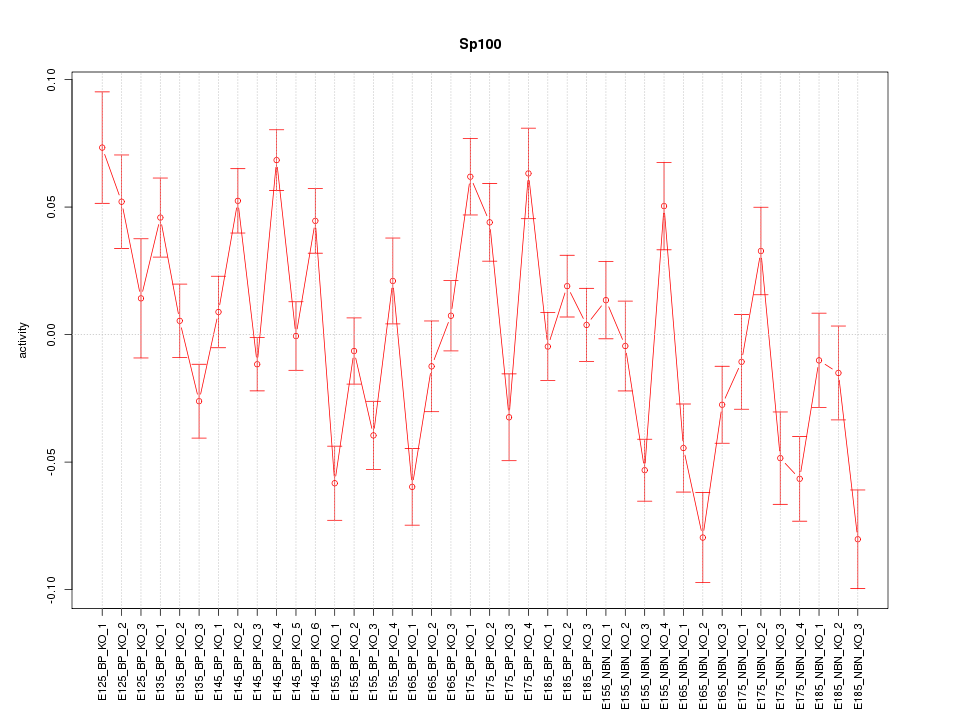
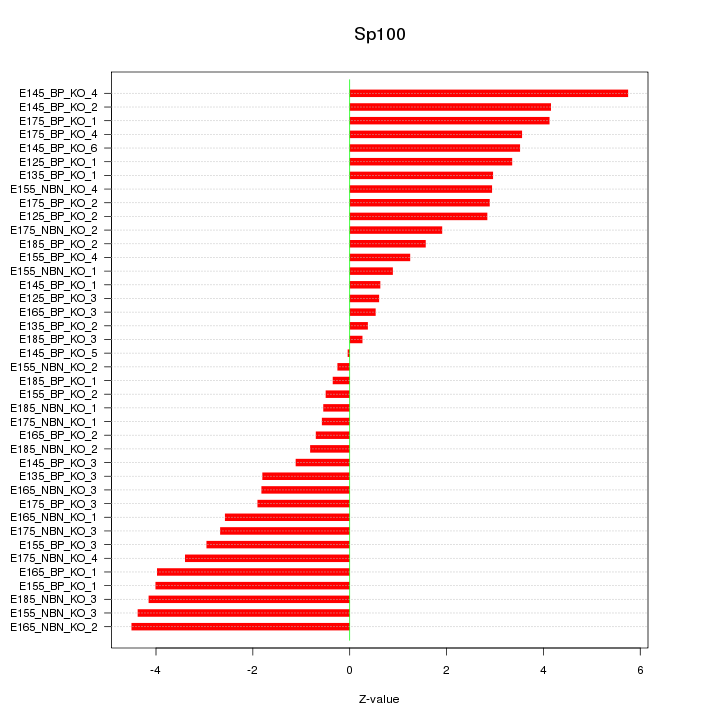
Motif ID: Sp100
Z-value: 2.663

Transcription factors associated with Sp100:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sp100 | ENSMUSG00000026222.10 | Sp100 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm10_v2_chr1_+_85650044_85650052 | 0.26 | 1.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 1.3 | 3.9 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.9 | 4.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.8 | 2.5 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.8 | 2.4 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.7 | 5.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.7 | 2.1 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.7 | 2.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.7 | 2.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.5 | 4.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.5 | 7.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.5 | 4.6 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.5 | 3.0 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.5 | 1.5 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.5 | 2.8 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.4 | 0.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 1.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 2.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 2.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 0.9 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 | 2.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 5.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.3 | 1.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.2 | 0.7 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 7.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 2.0 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 1.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 2.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 3.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.6 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 4.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 0.6 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 11.4 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.2 | 2.0 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.2 | 0.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 1.8 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 0.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 5.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 3.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.5 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 1.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 4.0 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 1.9 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.1 | 1.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.3 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.1 | 2.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 2.6 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 2.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 4.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 1.1 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.1 | 7.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.5 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.6 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 2.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 1.7 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 1.3 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 3.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 11.7 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.7 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.7 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 2.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 1.8 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 3.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.6 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0051103 | lagging strand elongation(GO:0006273) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.3 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.8 | 4.8 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 2.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.6 | 1.7 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.6 | 2.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.4 | 1.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.4 | 3.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 5.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 7.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 2.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 2.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 6.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 5.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 2.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 2.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 2.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.7 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 2.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 3.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 5.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 236.9 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.5 | 4.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.9 | 3.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.8 | 5.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 2.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.7 | 2.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 2.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.4 | 1.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 1.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 0.9 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.3 | 5.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 2.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 2.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 3.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 3.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 0.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 1.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.2 | 2.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 5.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 11.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 4.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 3.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 2.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.2 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 9.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 231.7 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 13.9 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 5.6 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.2 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.5 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.1 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 4.0 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.0 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.4 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 4.6 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 7.2 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.7 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.0 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 6.2 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.2 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.8 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.6 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 1.4 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 4.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.1 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 3.5 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.5 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.4 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME_BOTULINUM_NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.1 | REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 3.7 | REACTOME_DIABETES_PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.3 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 2.2 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |