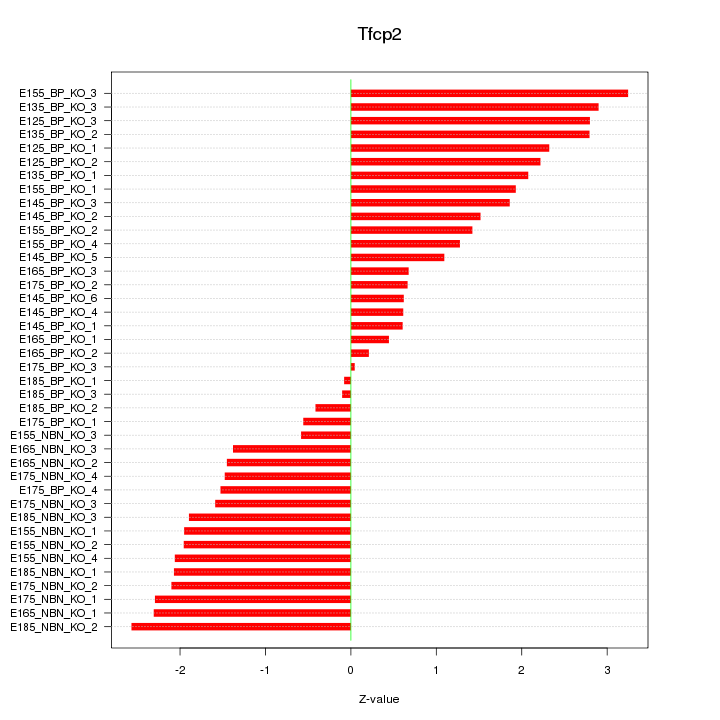
Motif ID: Tfcp2
Z-value: 1.725
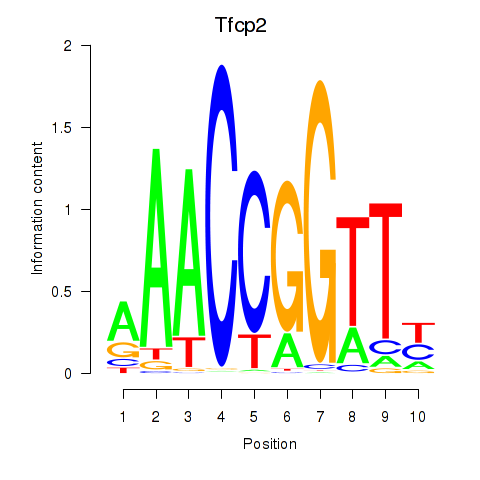
Transcription factors associated with Tfcp2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfcp2 | ENSMUSG00000009733.8 | Tfcp2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2 | mm10_v2_chr15_-_100551959_100552010 | 0.65 | 6.7e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.3 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 4.1 | 12.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 3.5 | 14.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 3.2 | 9.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 2.7 | 18.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 2.6 | 7.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 2.2 | 6.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.7 | 5.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.7 | 5.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 1.5 | 4.4 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.3 | 8.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) macropinocytosis(GO:0044351) |
| 1.2 | 5.9 | GO:0060266 | regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 1.2 | 5.9 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 1.2 | 3.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 1.1 | 3.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.1 | 8.8 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 1.1 | 5.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.9 | 3.6 | GO:1904778 | regulation of metaphase plate congression(GO:0090235) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.9 | 2.6 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.8 | 3.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.8 | 2.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.8 | 3.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.7 | 3.0 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.7 | 3.6 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.7 | 2.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.7 | 2.8 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.7 | 11.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.7 | 2.7 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.6 | 1.9 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.6 | 1.8 | GO:0038044 | negative regulation of receptor biosynthetic process(GO:0010871) transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 1.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 1.5 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.5 | 4.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 | 1.9 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.4 | 3.0 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 | 5.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 3.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.4 | 1.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 5.3 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 10.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.4 | 2.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.4 | 1.9 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 1.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 13.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 3.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 1.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 2.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 1.2 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.3 | 3.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 3.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 1.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.3 | 2.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 3.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 2.4 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.2 | 7.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 2.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.2 | 3.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.3 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.2 | 0.9 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) positive regulation of isotype switching to IgA isotypes(GO:0048298) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.9 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 | 1.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 1.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 4.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 3.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 3.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 1.7 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.2 | 1.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 2.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.5 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 1.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 2.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 4.2 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 8.1 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 4.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 2.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 2.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 16.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.6 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 1.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 9.8 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 4.0 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.1 | 3.1 | GO:0060042 | retina morphogenesis in camera-type eye(GO:0060042) |
| 0.1 | 0.8 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 1.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 2.2 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.3 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 4.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.0 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 1.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 5.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 3.6 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 3.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 2.7 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.0 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.4 | 9.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.8 | 5.3 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.2 | 5.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.0 | 7.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.9 | 3.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.9 | 2.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.7 | 3.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.7 | 2.8 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.7 | 3.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.6 | 1.8 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.6 | 2.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.6 | 2.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 3.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 3.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.5 | 4.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 3.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.4 | 5.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 8.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 3.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 2.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 5.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 0.9 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 18.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 2.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 17.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 9.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 17.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 6.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 8.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 4.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 3.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 8.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 5.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 2.8 | 14.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 2.5 | 12.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.4 | 9.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.9 | 17.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.7 | 5.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.5 | 4.4 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.2 | 5.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.1 | 17.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.1 | 13.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.1 | 3.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 1.0 | 3.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.9 | 3.5 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.7 | 3.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.6 | 2.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 1.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.5 | 7.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 1.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 5.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 1.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.4 | 4.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 2.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 1.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.4 | 3.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 16.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 2.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 3.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 3.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 2.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 0.9 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.3 | 0.9 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 3.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 3.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 2.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 2.4 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 1.6 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 2.3 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.2 | 0.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 3.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 9.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 1.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 1.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 6.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:2001070 | glycerophosphocholine phosphodiesterase activity(GO:0047389) starch binding(GO:2001070) |
| 0.1 | 0.8 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 6.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 3.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 4.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.5 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.5 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 5.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 3.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 2.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 3.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 3.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 12.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 4.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 4.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) pyrroline-5-carboxylate reductase activity(GO:0004735) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 17.3 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.5 | 23.5 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.3 | 12.8 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.3 | 13.6 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.2 | 5.3 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 6.6 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 14.9 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 4.6 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.6 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 1.8 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 15.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.7 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.1 | 1.6 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.7 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.9 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.4 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.7 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.5 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 3.2 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.6 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 17.3 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 1.3 | 13.1 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 1.2 | 18.8 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 9.5 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.5 | 7.0 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 5.1 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 5.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.3 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 5.1 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 3.0 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 12.4 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 5.0 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 2.1 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.8 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.6 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.0 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.3 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.0 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.9 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 6.9 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.7 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.7 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.3 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 2.2 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.5 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 3.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 14.9 | REACTOME_HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 3.8 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.8 | REACTOME_DNA_REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.6 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.3 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 3.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.2 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |