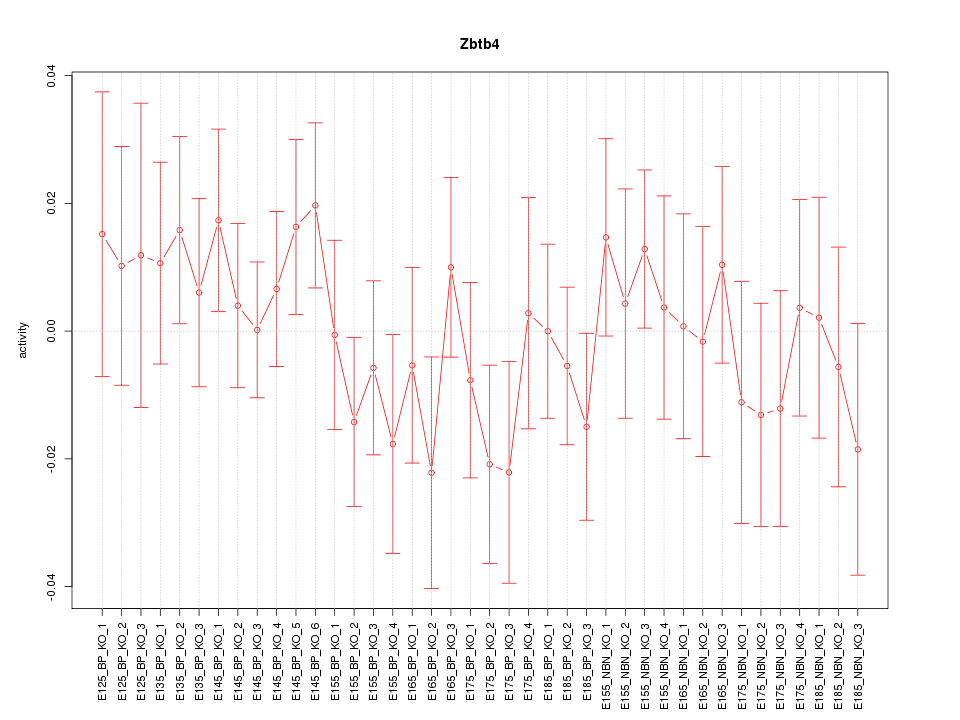
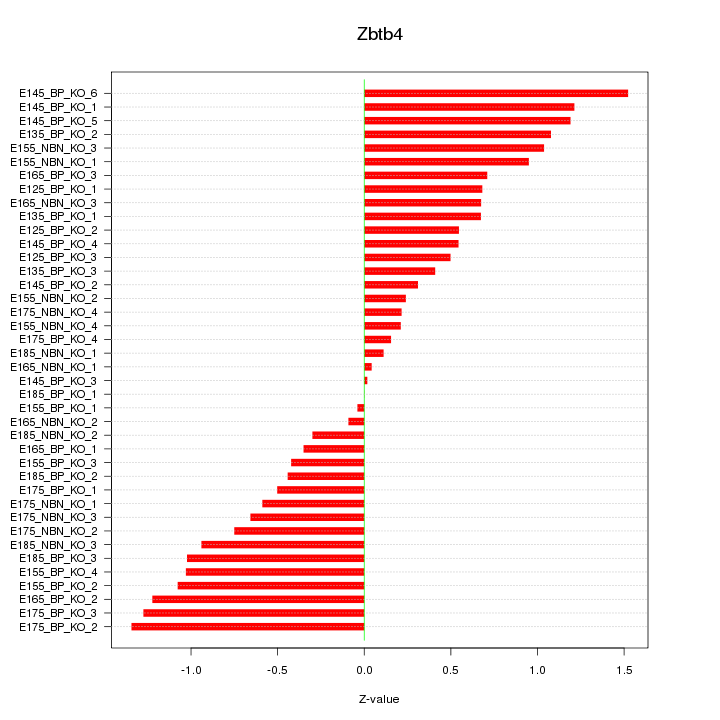
Motif ID: Zbtb4
Z-value: 0.755

Transcription factors associated with Zbtb4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb4 | ENSMUSG00000018750.8 | Zbtb4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | mm10_v2_chr11_+_69765899_69765925 | -0.37 | 1.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.0 | 7.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.9 | 4.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.7 | 5.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 2.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 0.8 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.4 | 3.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.4 | 1.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.6 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.3 | 0.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.3 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.7 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 1.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 0.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 2.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 1.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 0.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.8 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.9 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.9 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.4 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.3 | GO:0051794 | positive regulation of keratinocyte proliferation(GO:0010838) regulation of catagen(GO:0051794) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.2 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 2.2 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.8 | GO:0071436 | sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 2.0 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.5 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.4 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 0.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.3 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 | 1.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.7 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.6 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.4 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.3 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0060973 | cell migration involved in heart development(GO:0060973) regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.8 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.9 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.5 | GO:0003341 | cilium movement(GO:0003341) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 4.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 3.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 0.5 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 2.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.4 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.9 | GO:0099568 | cytoplasmic region(GO:0099568) |
| 0.1 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.4 | 2.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.4 | 2.5 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.4 | 1.6 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.3 | 1.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 0.9 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.3 | 4.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 2.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 3.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.8 | GO:0050786 | arachidonic acid binding(GO:0050544) RAGE receptor binding(GO:0050786) |
| 0.1 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.8 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 3.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.7 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 1.7 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.2 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.1 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.8 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 1.5 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.7 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.1 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.9 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.1 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.6 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.1 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |