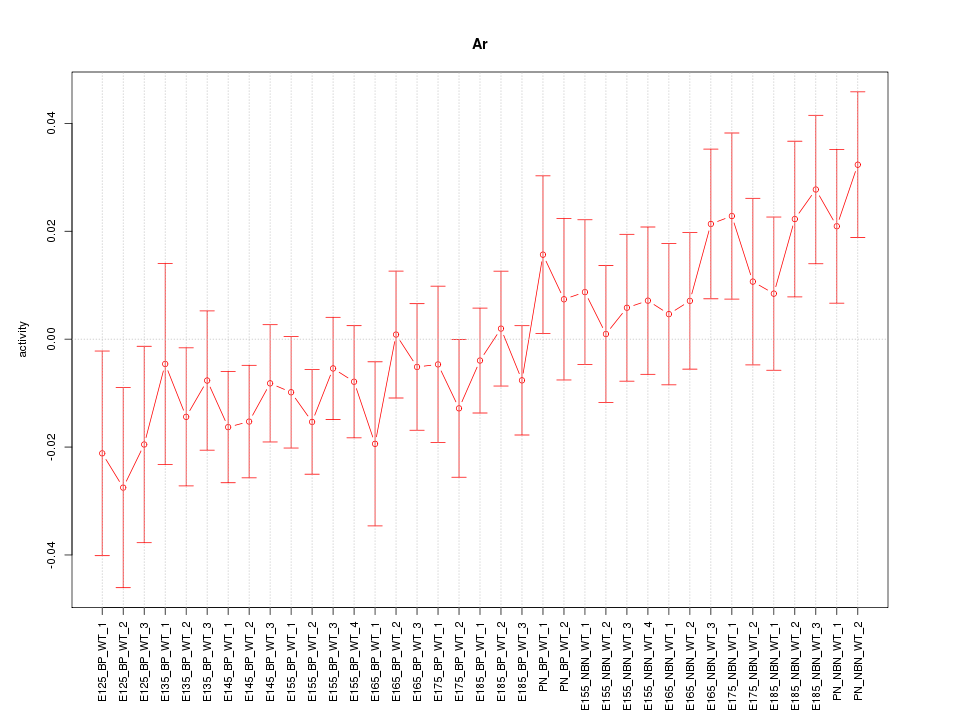
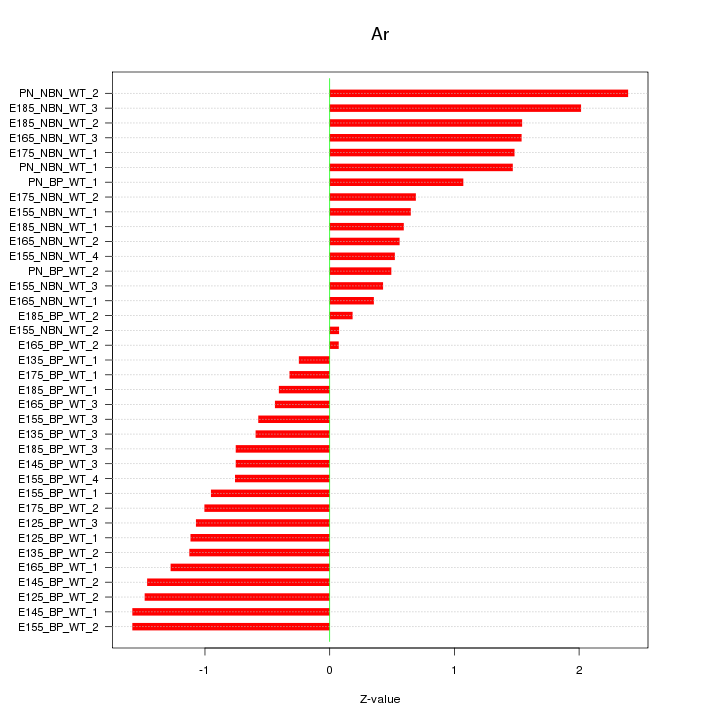
Motif ID: Ar
Z-value: 1.065
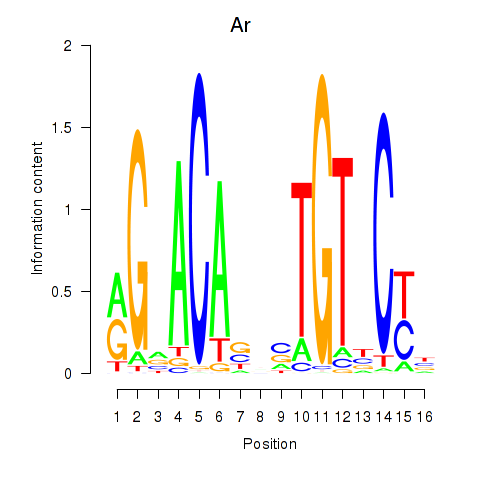
Transcription factors associated with Ar:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ar | ENSMUSG00000046532.7 | Ar |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.3 | 3.9 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.1 | 12.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.0 | 3.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.9 | 6.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.8 | 4.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.8 | 2.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.8 | 3.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.8 | 2.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.7 | 2.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.7 | 4.6 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.6 | 1.9 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.6 | 2.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 1.6 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.5 | 2.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.5 | 1.4 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.5 | 2.3 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.4 | 1.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.4 | 2.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 1.6 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.4 | 2.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.4 | 1.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 0.7 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.3 | 1.0 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 0.6 | GO:0002481 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.3 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.5 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.3 | 0.8 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.3 | 1.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 1.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 1.5 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 1.0 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 0.6 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 0.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.3 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.2 | 0.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 2.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 2.5 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.2 | 2.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 0.8 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 0.9 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.8 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 2.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 3.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.0 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.6 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.1 | 0.4 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.6 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 1.2 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 9.7 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 2.0 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 0.1 | 1.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.1 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.1 | 0.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.9 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 1.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 1.7 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 2.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.6 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 1.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 1.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 2.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 0.1 | GO:1903061 | glucosylceramide catabolic process(GO:0006680) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 1.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.2 | 6.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 2.0 | GO:0097487 | vesicle lumen(GO:0031983) multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 1.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.4 | 1.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 1.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 3.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 0.7 | GO:0002141 | stereocilia ankle link(GO:0002141) |
| 0.2 | 1.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 1.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 11.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 2.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 1.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 10.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 11.9 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 2.0 | 6.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.3 | 3.9 | GO:0031403 | lithium ion binding(GO:0031403) |
| 1.0 | 3.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.8 | 2.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.8 | 3.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.8 | 3.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 12.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.7 | 2.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 2.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.4 | 1.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.4 | 2.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 2.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.3 | 2.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.0 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 2.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.2 | 2.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.2 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 2.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.5 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 2.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 3.2 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.0 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.0 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.2 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 8.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.8 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.9 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.8 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 1.4 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.8 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.0 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.2 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.2 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.8 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.1 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 3.0 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.4 | 3.0 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 3.8 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.1 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.9 | REACTOME_GRB2_EVENTS_IN_ERBB2_SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 1.4 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.2 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 5.7 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.9 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 10.4 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.5 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 3.0 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.8 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.8 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.0 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.8 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.9 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.3 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |