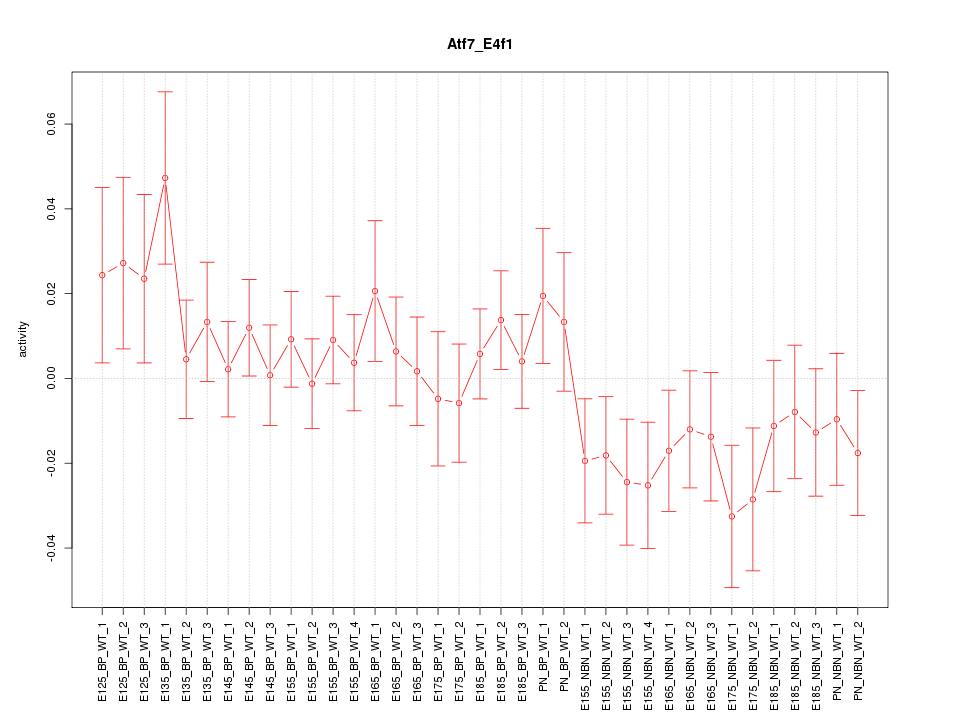
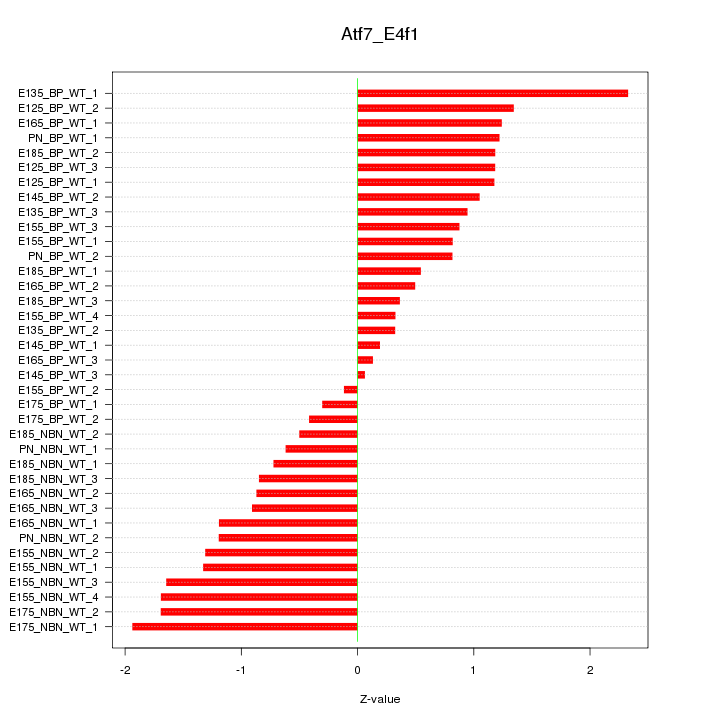
Motif ID: Atf7_E4f1
Z-value: 1.062
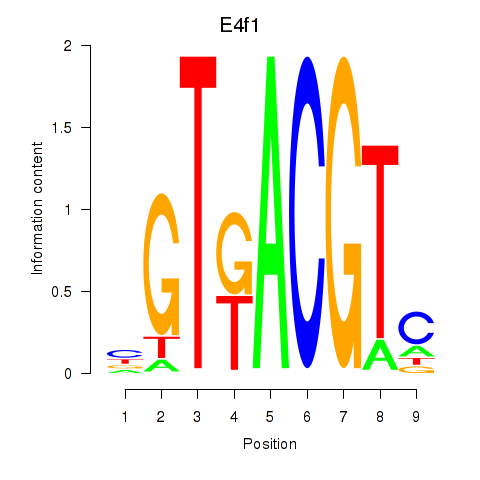
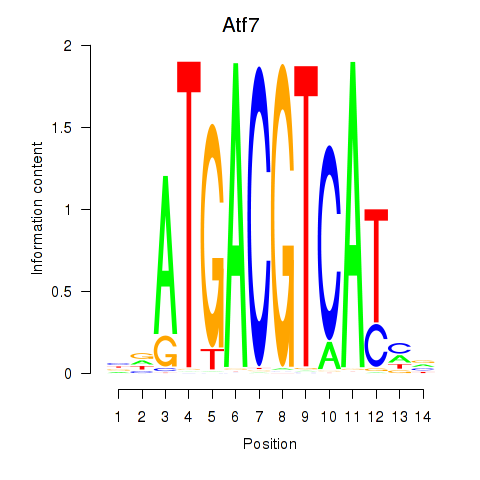
Transcription factors associated with Atf7_E4f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atf7 | ENSMUSG00000052414.9 | Atf7 |
| Atf7 | ENSMUSG00000071584.1 | Atf7 |
| E4f1 | ENSMUSG00000024137.8 | E4f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E4f1 | mm10_v2_chr17_-_24455265_24455345 | -0.14 | 4.2e-01 | Click! |
| Atf7 | mm10_v2_chr15_-_102529025_102529025 | -0.13 | 4.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.2 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.2 | 8.8 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 2.2 | 6.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.8 | 21.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 1.4 | 4.3 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 1.2 | 7.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.2 | 3.5 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 1.1 | 3.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.1 | 5.4 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 1.0 | 7.2 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 1.0 | 5.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 1.0 | 10.6 | GO:0030432 | peristalsis(GO:0030432) |
| 0.9 | 6.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.9 | 2.8 | GO:0072284 | cervix development(GO:0060067) metanephric S-shaped body morphogenesis(GO:0072284) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.7 | 2.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.7 | 6.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.7 | 2.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 2.4 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.6 | 5.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.6 | 5.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.6 | 2.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.6 | 1.7 | GO:1901079 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) |
| 0.5 | 1.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.5 | 1.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.5 | 7.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.5 | 5.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.4 | 2.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.4 | 2.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 3.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.4 | 1.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 0.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.3 | 3.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.3 | 6.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.3 | 1.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.3 | 1.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.3 | 3.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 | 1.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 1.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.3 | 1.3 | GO:0060578 | extraocular skeletal muscle development(GO:0002074) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 3.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 0.9 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 | 0.9 | GO:1902527 | regulation of protein monoubiquitination(GO:1902525) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.3 | 0.9 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.3 | 3.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 1.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 8.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 3.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 3.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 8.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.3 | 2.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 4.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.3 | 1.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 0.8 | GO:2001016 | heparan sulfate proteoglycan catabolic process(GO:0030200) positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.2 | 3.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 1.2 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.2 | 0.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 1.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.2 | 1.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 7.9 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 0.7 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 0.9 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.8 | GO:0032202 | negative regulation of protein ADP-ribosylation(GO:0010836) telomere assembly(GO:0032202) |
| 0.2 | 0.8 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 1.0 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 3.6 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 2.8 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.2 | 1.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.7 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.2 | 3.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 1.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 1.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.4 | GO:0045658 | eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.7 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.0 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.7 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 2.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.6 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.7 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 1.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 4.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 2.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.6 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.1 | 0.6 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 2.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.3 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.8 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 2.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.8 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.2 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) modulation by virus of host autophagy(GO:0039519) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 0.5 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.9 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 3.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.6 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 1.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 3.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 2.9 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of selenocysteine incorporation(GO:1904569) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.3 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 3.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.0 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.2 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.0 | 0.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.2 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0016569 | covalent chromatin modification(GO:0016569) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.4 | 13.7 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.2 | 8.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.2 | 4.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.2 | 7.2 | GO:0098536 | deuterosome(GO:0098536) |
| 1.2 | 3.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 1.1 | 3.3 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.9 | 6.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.7 | 6.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 3.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.5 | 1.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.5 | 3.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 2.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 5.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 2.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 1.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 1.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 4.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.3 | 1.8 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 0.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 8.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 5.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 1.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 5.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 7.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 8.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 1.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.4 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 11.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 16.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.3 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.1 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 7.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 3.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 3.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 4.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 5.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.5 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 2.0 | 10.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.9 | 8.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.8 | 13.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 2.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.6 | 9.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 2.3 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.6 | 3.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.5 | 1.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 2.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 2.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 1.8 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.4 | 2.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 3.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 2.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 5.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 1.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 8.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 1.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.3 | 2.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.3 | 2.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 3.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 4.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 7.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 0.7 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.2 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 10.6 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 0.6 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 0.8 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 2.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.5 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 2.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 3.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 4.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.7 | GO:0070180 | ligase inhibitor activity(GO:0055104) large ribosomal subunit rRNA binding(GO:0070180) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 18.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 3.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 3.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 3.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 2.5 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 2.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 2.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 2.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 18.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.2 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.5 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.6 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 2.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 2.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 10.0 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.3 | 12.9 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.3 | 15.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.2 | 8.5 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 2.3 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 0.6 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 3.2 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.7 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 7.1 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 1.3 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.8 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 5.9 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.0 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 3.1 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 1.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID_HIV_NEF_PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.2 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.8 | 6.5 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.7 | 8.8 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.7 | 6.6 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 8.6 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 12.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.3 | 1.2 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.0 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 12.7 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.3 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 1.7 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 4.5 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 0.8 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 4.9 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.2 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 9.2 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.5 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 5.8 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 6.9 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 8.5 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.0 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.3 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_MULTIPLE_NUCLEOTIDE_PATCH_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.1 | 2.5 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.7 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.4 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.5 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 3.2 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.6 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 3.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.8 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.3 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.0 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.4 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.9 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.4 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.7 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |