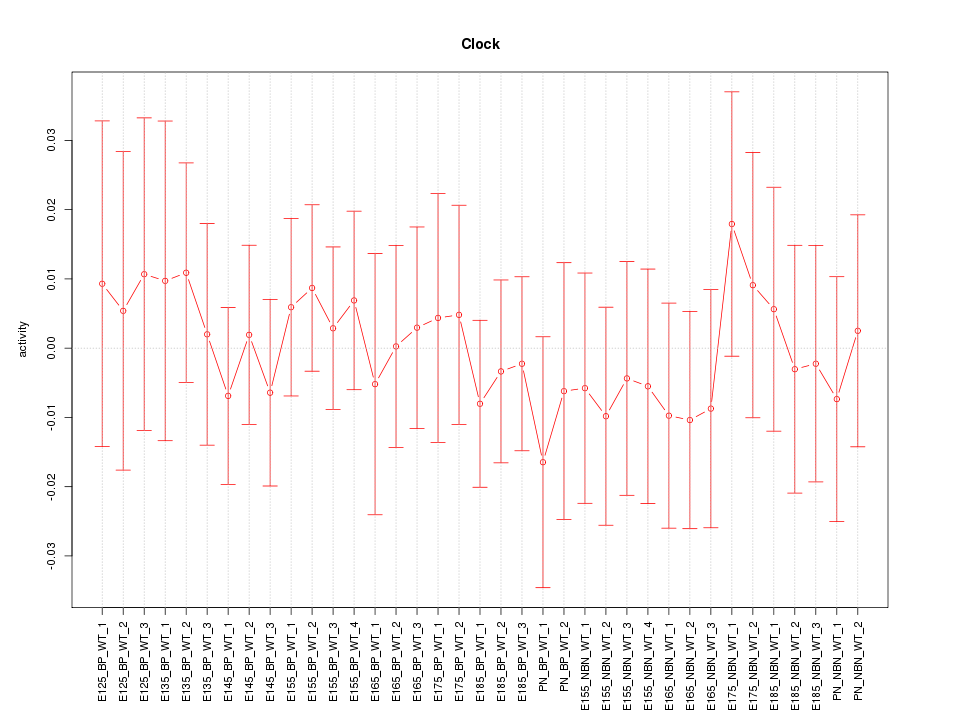
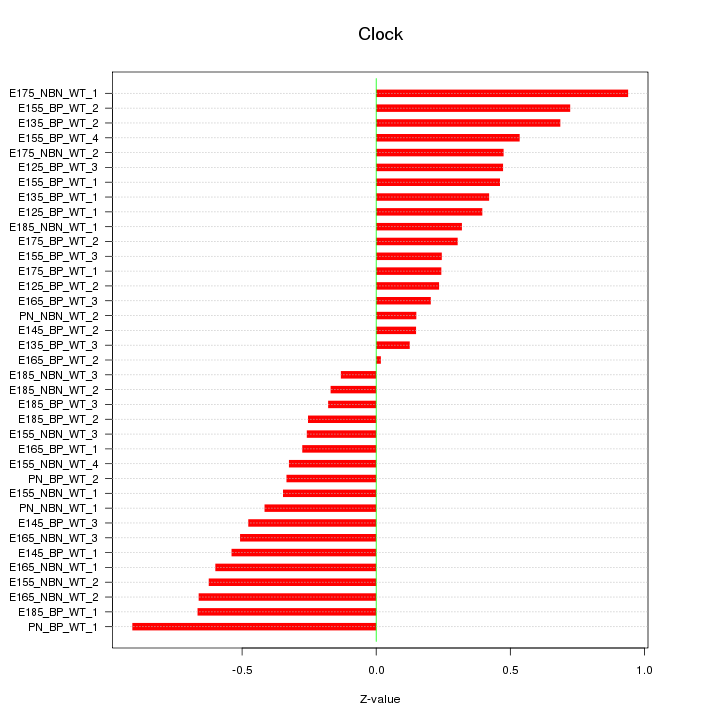
Motif ID: Clock
Z-value: 0.455
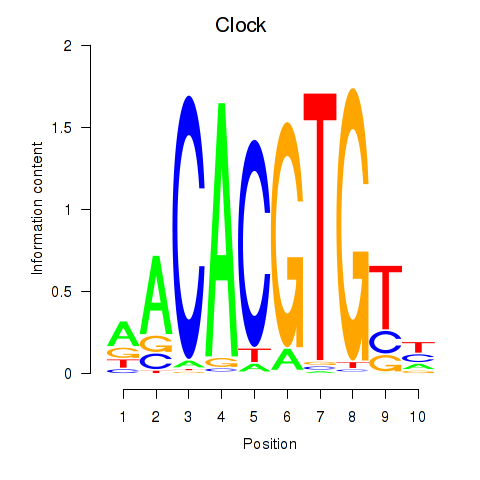
Transcription factors associated with Clock:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Clock | ENSMUSG00000029238.8 | Clock |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Clock | mm10_v2_chr5_-_76304474_76304548 | 0.16 | 3.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 0.7 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 0.6 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.2 | 1.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 0.7 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.2 | 0.7 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.2 | 1.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 1.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 1.0 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.6 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.3 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.3 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.1 | 0.6 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.5 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0097278 | transforming growth factor beta activation(GO:0036363) complement-dependent cytotoxicity(GO:0097278) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.4 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.4 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 | 0.2 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.2 | GO:0032310 | drug transmembrane transport(GO:0006855) prostaglandin transport(GO:0015732) prostaglandin secretion(GO:0032310) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.4 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 0.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 0.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.7 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.6 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.0 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.1 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.1 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.0 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.8 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME_TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.1 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |