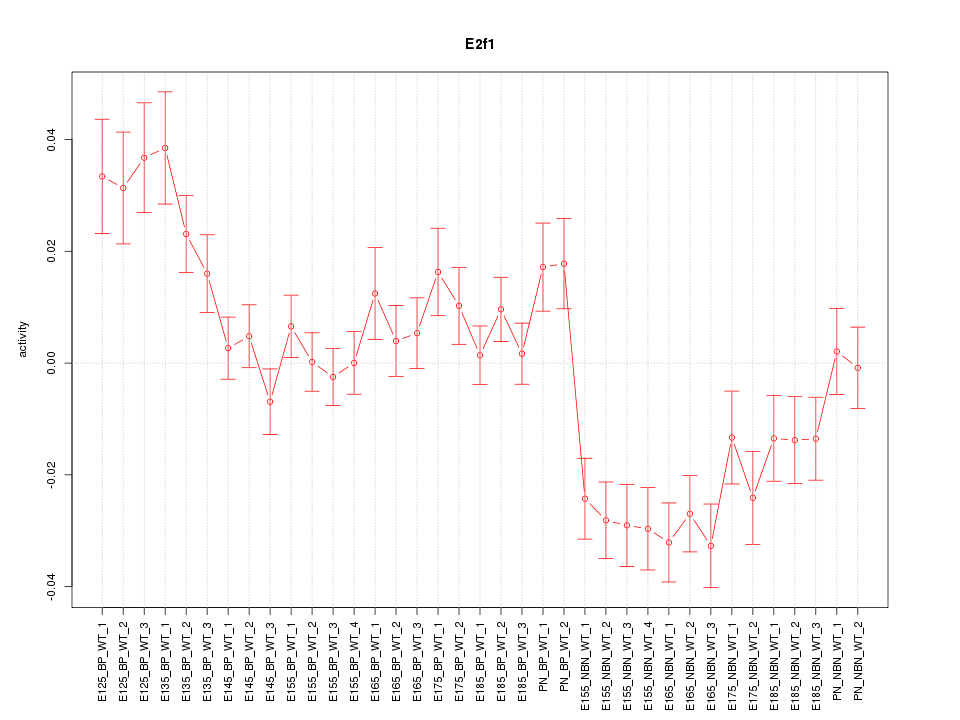
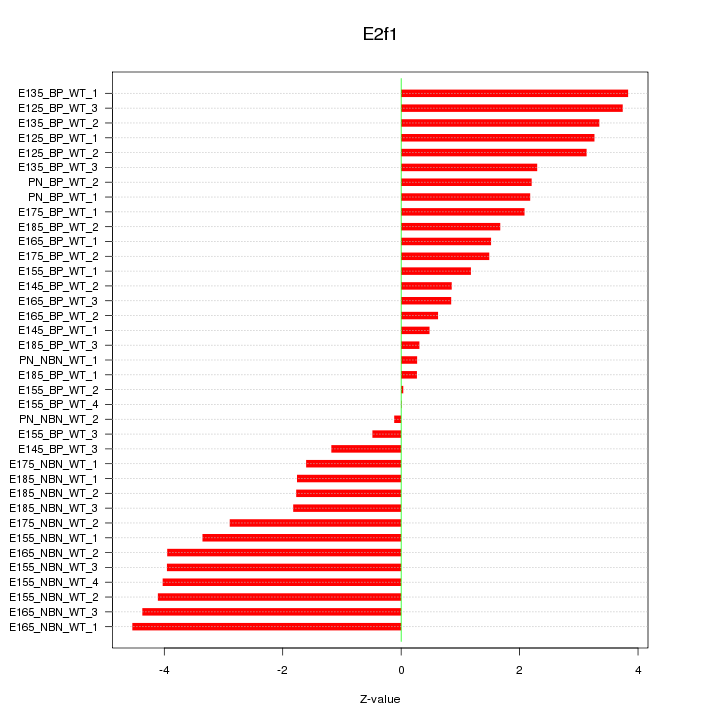
Motif ID: E2f1
Z-value: 2.483
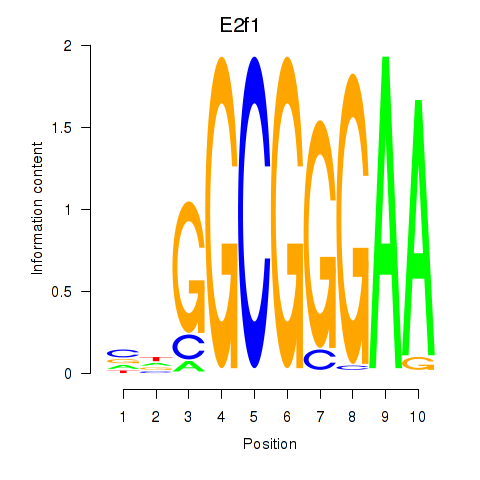
Transcription factors associated with E2f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f1 | ENSMUSG00000027490.11 | E2f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f1 | mm10_v2_chr2_-_154569720_154569799 | 0.90 | 1.7e-14 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 24.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 4.3 | 21.5 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 3.6 | 10.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 3.5 | 10.4 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 3.2 | 9.6 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 3.1 | 9.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 2.9 | 11.6 | GO:2000768 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 2.7 | 18.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 2.6 | 7.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 2.5 | 2.5 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 2.5 | 10.1 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 2.4 | 36.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 2.4 | 7.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.4 | 4.8 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 2.3 | 22.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 2.2 | 11.1 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 2.1 | 8.6 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 2.0 | 18.1 | GO:0033504 | floor plate development(GO:0033504) |
| 2.0 | 6.0 | GO:0003360 | brainstem development(GO:0003360) |
| 2.0 | 29.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.9 | 7.7 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.9 | 1.9 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 1.8 | 20.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 1.8 | 7.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 1.7 | 25.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 1.7 | 8.5 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 1.7 | 1.7 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 1.7 | 8.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 1.6 | 4.8 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.5 | 6.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 1.5 | 7.6 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 1.5 | 26.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.4 | 5.7 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.4 | 4.3 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.4 | 4.1 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 1.4 | 5.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.4 | 21.7 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 1.3 | 4.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 1.3 | 3.9 | GO:0090425 | hepatocyte cell migration(GO:0002194) pancreas morphogenesis(GO:0061113) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.3 | 27.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 1.3 | 6.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.2 | 8.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.2 | 10.8 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.2 | 6.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.2 | 5.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 1.2 | 22.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.2 | 7.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.2 | 14.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 1.1 | 3.4 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.1 | 4.6 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.1 | 4.5 | GO:0010288 | response to lead ion(GO:0010288) |
| 1.1 | 1.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.1 | 3.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.0 | 12.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 1.0 | 6.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 1.0 | 19.1 | GO:0007530 | sex determination(GO:0007530) |
| 1.0 | 3.0 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 1.0 | 2.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 1.0 | 5.8 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 1.0 | 6.7 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.9 | 4.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.9 | 3.7 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.9 | 2.7 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.9 | 2.7 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.9 | 6.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.8 | 22.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.8 | 5.7 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.8 | 4.0 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.8 | 8.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.8 | 14.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.8 | 4.8 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.8 | 1.6 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.8 | 0.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.8 | 1.6 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.8 | 3.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.8 | 2.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.7 | 5.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 2.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.7 | 1.4 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.7 | 2.8 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.7 | 4.9 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.7 | 6.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.7 | 2.0 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.7 | 10.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.6 | 1.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.6 | 1.9 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.6 | 1.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.6 | 3.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.6 | 6.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.6 | 6.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.6 | 6.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.6 | 5.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.6 | 1.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.6 | 2.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.6 | 2.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.5 | 2.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 1.6 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.5 | 1.6 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.5 | 3.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.5 | 2.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.5 | 2.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.5 | 11.5 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.5 | 11.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.5 | 5.0 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.5 | 1.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.5 | 11.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.5 | 1.9 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.5 | 1.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.5 | 4.3 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.5 | 1.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.5 | 2.8 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 0.9 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.5 | 1.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 1.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.5 | 3.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.5 | 1.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.4 | 0.9 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.4 | 4.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 0.4 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.4 | 0.9 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.4 | 1.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.4 | 5.1 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.4 | 3.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.4 | 5.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 2.9 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.4 | 1.6 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.4 | 2.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.4 | 3.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 2.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.4 | 1.6 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.4 | 1.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.4 | 3.6 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.4 | 9.5 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.4 | 6.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 1.1 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 0.4 | 1.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 1.9 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.4 | 1.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.4 | 3.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.4 | 1.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.4 | 3.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.3 | 2.8 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 2.4 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 2.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 1.0 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.3 | 3.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.3 | 0.3 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.3 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 1.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.3 | 7.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.3 | 3.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 1.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.3 | 5.7 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.3 | 7.5 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.3 | 4.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 2.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 0.9 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 0.9 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 0.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.3 | 5.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.3 | 10.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.3 | 10.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.3 | 6.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 0.6 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.3 | 1.4 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.3 | 0.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.3 | 1.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 7.1 | GO:0046605 | regulation of centrosome cycle(GO:0046605) |
| 0.3 | 2.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 2.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 2.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.3 | 2.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 2.8 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.3 | 1.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 1.1 | GO:0051562 | cellular triglyceride homeostasis(GO:0035356) negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 2.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 0.5 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.3 | 0.8 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 0.3 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.3 | 0.8 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.3 | 8.0 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.3 | 6.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.3 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.3 | 4.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 0.8 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 0.5 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.2 | 1.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 0.7 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.2 | 2.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 0.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 5.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 1.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 0.5 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.2 | 2.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.2 | 0.2 | GO:0072069 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.2 | 6.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.2 | 2.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.9 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 1.6 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 3.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 9.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.2 | 0.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 2.0 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.9 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.2 | 0.6 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.2 | 4.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 5.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.2 | 1.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 1.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.2 | 0.8 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.2 | 0.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 4.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 0.6 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 2.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 0.8 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 1.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 3.0 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 0.9 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 1.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 0.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 0.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.5 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.2 | 1.6 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 | 0.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 | 2.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 0.7 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.2 | 0.8 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.2 | 0.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 0.2 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.2 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 5.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 1.0 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.2 | 1.1 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.2 | 1.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.2 | 0.8 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 0.5 | GO:0001907 | killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.2 | 2.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 0.5 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.2 | 1.4 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 0.9 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.7 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.4 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 2.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 2.1 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.7 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 6.0 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.6 | GO:1990776 | angiotensin-activated signaling pathway(GO:0038166) cellular response to angiotensin(GO:1904385) response to angiotensin(GO:1990776) |
| 0.1 | 1.5 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.4 | GO:0060430 | lung saccule development(GO:0060430) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 3.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.0 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 4.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 3.9 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 1.2 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.4 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.1 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 4.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.7 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.1 | 1.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.0 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.1 | 0.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.6 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) specification of loop of Henle identity(GO:0072086) |
| 0.1 | 3.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 3.7 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 0.3 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.1 | 0.9 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 2.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 5.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.4 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:0043987 | histone-serine phosphorylation(GO:0035404) histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 0.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 9.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 2.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.6 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 2.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 3.6 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 0.7 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.9 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.6 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 1.3 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 | 3.1 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 1.3 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 2.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.4 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.0 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.3 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.3 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 4.8 | 23.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 3.6 | 10.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.9 | 28.7 | GO:0000796 | condensin complex(GO:0000796) |
| 2.8 | 16.6 | GO:0098536 | deuterosome(GO:0098536) |
| 2.7 | 10.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 2.4 | 7.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.2 | 11.0 | GO:0031523 | Myb complex(GO:0031523) |
| 2.1 | 6.3 | GO:0035101 | FACT complex(GO:0035101) |
| 2.1 | 10.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 2.0 | 6.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.9 | 5.7 | GO:0071914 | prominosome(GO:0071914) |
| 1.7 | 6.9 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.6 | 4.9 | GO:0000801 | central element(GO:0000801) |
| 1.6 | 4.8 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.4 | 5.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.4 | 5.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.3 | 7.9 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.2 | 7.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.1 | 13.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.1 | 10.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.1 | 4.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.0 | 10.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.0 | 4.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.9 | 30.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.9 | 2.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.8 | 3.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.8 | 2.4 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.8 | 3.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.8 | 58.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.8 | 3.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.7 | 4.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.7 | 2.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.7 | 1.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.7 | 3.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.7 | 4.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.7 | 2.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.7 | 11.3 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.6 | 3.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.6 | 3.0 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.6 | 3.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.6 | 3.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.6 | 2.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.6 | 1.7 | GO:0071920 | cleavage body(GO:0071920) |
| 0.5 | 1.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.5 | 3.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 1.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.5 | 8.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 8.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 3.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 3.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.5 | 4.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 4.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 4.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.4 | 2.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.4 | 2.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.4 | 2.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 2.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 5.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 1.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 2.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 5.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 2.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 2.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 4.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 1.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 4.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 1.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 4.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 0.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 6.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 2.4 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 3.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 6.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 3.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 1.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.3 | 4.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 4.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 25.5 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 2.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.3 | 2.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 1.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 42.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.3 | 2.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 4.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 0.6 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.2 | 1.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 2.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 9.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 3.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 0.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 6.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 4.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 5.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 0.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 2.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 1.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.5 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 46.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 3.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.9 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 5.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 0.8 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 6.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 2.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 24.0 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.1 | 2.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 12.8 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.9 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 6.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 5.5 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 43.2 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 3.0 | GO:0005924 | cell-substrate adherens junction(GO:0005924) cell-substrate junction(GO:0030055) |
| 0.0 | 0.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 38.3 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 29.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 4.8 | 23.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 4.0 | 4.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 3.9 | 11.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 3.7 | 22.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 2.4 | 12.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.2 | 11.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.9 | 13.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.9 | 5.7 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 1.8 | 9.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.8 | 1.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.6 | 4.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.5 | 7.7 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 1.5 | 4.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.5 | 7.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.3 | 7.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.2 | 9.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.2 | 7.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.2 | 4.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.1 | 3.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 1.1 | 12.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.0 | 6.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.9 | 3.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.9 | 2.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.9 | 5.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.9 | 13.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 3.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.8 | 6.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.8 | 7.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.8 | 4.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.8 | 5.4 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.7 | 5.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.7 | 2.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.7 | 3.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.7 | 7.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.7 | 4.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.7 | 7.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.6 | 1.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 3.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.6 | 1.9 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.6 | 3.8 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.6 | 2.3 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.6 | 3.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.6 | 20.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.6 | 3.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.5 | 12.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 14.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.5 | 1.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.5 | 24.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.5 | 7.5 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.5 | 6.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.5 | 2.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.5 | 5.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.4 | 3.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 1.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.4 | 17.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.4 | 4.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 1.6 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.4 | 0.8 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.4 | 1.9 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.4 | 2.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.4 | 4.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 1.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.4 | 1.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.4 | 3.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 1.0 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 4.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 1.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 1.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 7.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 0.9 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 7.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 1.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 3.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.5 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 3.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.3 | 3.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 1.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 0.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.3 | 0.6 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 1.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 1.1 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.3 | 2.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 4.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 2.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 56.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 1.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 2.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 1.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 3.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 10.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 3.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 0.7 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.2 | 0.9 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 0.7 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.2 | 2.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 4.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 1.1 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.2 | 2.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 0.8 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 7.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 25.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 7.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 4.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 1.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 1.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 3.0 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 32.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 0.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 7.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 1.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.8 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 3.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 0.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.1 | 0.4 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.6 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 8.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 5.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 9.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 2.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 3.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 5.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 2.6 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 3.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 9.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 12.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 7.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 10.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.4 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 4.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.2 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 2.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 10.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0044020 | snoRNP binding(GO:0030519) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.1 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 55.2 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 5.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 9.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.6 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 1.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 3.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 4.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 22.8 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.2 | 10.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.2 | 55.3 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 1.0 | 80.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.8 | 31.7 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.6 | 19.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 1.7 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 8.6 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.3 | 3.8 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 9.9 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 4.0 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.2 | 3.9 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 4.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.2 | 5.6 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 10.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.3 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 7.1 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 14.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 5.3 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.2 | 3.6 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 2.1 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 2.0 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.2 | 3.6 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 6.0 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.2 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.1 | 4.2 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.1 | 4.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.2 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 4.8 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.9 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 3.1 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 0.6 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.5 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 0.7 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.3 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.5 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 5.2 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.5 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.6 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.9 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 6.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.2 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 0.9 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.9 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID_LYMPH_ANGIOGENESIS_PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.2 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 33.3 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 1.8 | 36.0 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.4 | 2.8 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.2 | 8.4 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 10.9 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.0 | 4.8 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.9 | 11.6 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.8 | 5.9 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.8 | 3.3 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |
| 0.8 | 44.4 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.8 | 15.1 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.8 | 6.8 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.7 | 9.4 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.6 | 8.6 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.6 | 4.5 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.5 | 14.8 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.5 | 11.6 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.5 | 11.6 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 11.5 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 14.5 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 4.2 | REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.3 | 3.8 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 3.6 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 10.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.3 | 12.4 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.3 | 23.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 1.3 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 15.9 | REACTOME_RECRUITMENT_OF_MITOTIC_CENTROSOME_PROTEINS_AND_COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.2 | 4.7 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.2 | 4.1 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.6 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 7.0 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 0.9 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 3.6 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 10.5 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 1.4 | REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.2 | 0.6 | REACTOME_MITOTIC_G2_G2_M_PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.2 | 1.4 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 1.1 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 4.3 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 1.6 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.2 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.0 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 5.0 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.5 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.7 | REACTOME_RNA_POL_I_TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 2.5 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.8 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.9 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.8 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.6 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.1 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.8 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.0 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.7 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.8 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.1 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.6 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 2.7 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 0.4 | REACTOME_SHC_MEDIATED_SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 0.3 | REACTOME_DNA_STRAND_ELONGATION | Genes involved in DNA strand elongation |
| 0.1 | 2.6 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.3 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.3 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.0 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 2.4 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 19.3 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.6 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.8 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.4 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.8 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.8 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.2 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.7 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.4 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.2 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.7 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.5 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.9 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.0 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 2.7 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.0 | REACTOME_APOPTOTIC_EXECUTION_PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.1 | REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME_INTERFERON_SIGNALING | Genes involved in Interferon Signaling |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |