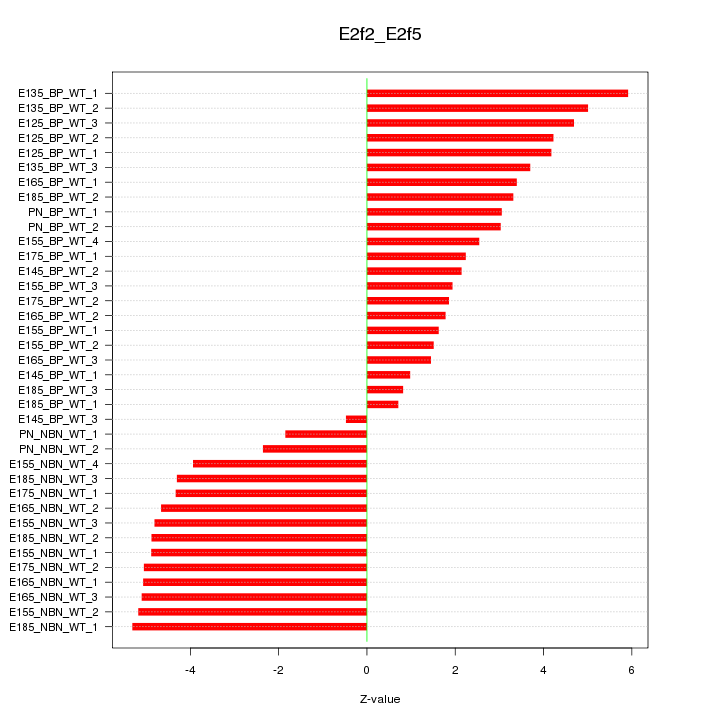
Motif ID: E2f2_E2f5
Z-value: 3.657
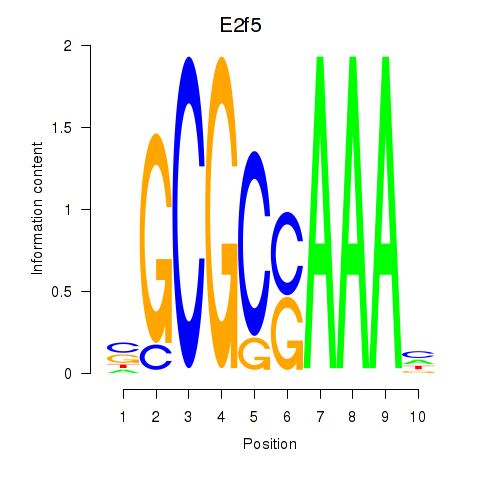
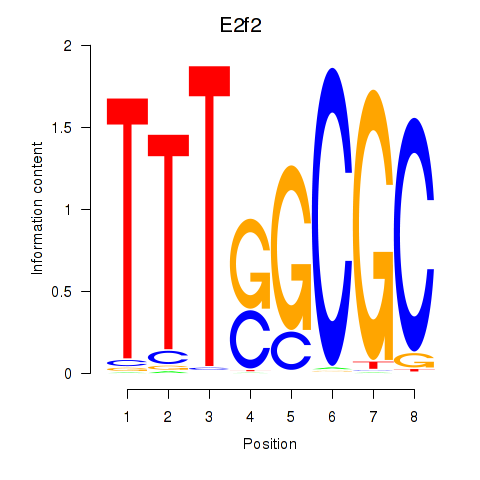
Transcription factors associated with E2f2_E2f5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f2 | ENSMUSG00000018983.9 | E2f2 |
| E2f5 | ENSMUSG00000027552.8 | E2f5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f2 | mm10_v2_chr4_+_136172367_136172395 | 0.95 | 6.3e-19 | Click! |
| E2f5 | mm10_v2_chr3_+_14578609_14578687 | 0.79 | 6.6e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.8 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 9.1 | 63.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 6.9 | 55.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 6.8 | 67.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 5.0 | 15.0 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 4.8 | 43.1 | GO:0033504 | floor plate development(GO:0033504) |
| 4.8 | 4.8 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 4.7 | 42.0 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 4.5 | 18.0 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 4.3 | 25.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 3.7 | 14.8 | GO:0046655 | glycine biosynthetic process(GO:0006545) folic acid metabolic process(GO:0046655) |
| 3.6 | 14.5 | GO:0006272 | leading strand elongation(GO:0006272) |
| 3.3 | 16.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 3.2 | 45.0 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 2.8 | 48.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 2.8 | 8.4 | GO:1904347 | intestine smooth muscle contraction(GO:0014827) regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) regulation of small intestine smooth muscle contraction(GO:1904347) small intestine smooth muscle contraction(GO:1990770) |
| 2.7 | 24.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.6 | 7.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 2.5 | 7.5 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 2.5 | 69.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 2.1 | 10.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.9 | 7.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 1.9 | 5.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.7 | 12.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 1.7 | 5.2 | GO:0072554 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.7 | 16.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.6 | 8.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 1.6 | 8.0 | GO:0019230 | proprioception(GO:0019230) |
| 1.5 | 9.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 1.5 | 5.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 1.4 | 8.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 1.4 | 4.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 1.3 | 5.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.3 | 15.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.3 | 3.8 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 1.2 | 5.0 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.2 | 12.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.2 | 8.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.1 | 9.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.1 | 17.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 1.1 | 4.3 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 1.1 | 4.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.0 | 56.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 1.0 | 15.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.0 | 3.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.0 | 14.3 | GO:0072189 | ureter development(GO:0072189) |
| 1.0 | 3.0 | GO:0060423 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 1.0 | 4.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.9 | 0.9 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.9 | 4.3 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.9 | 8.7 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.8 | 3.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.8 | 4.9 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.8 | 7.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.8 | 36.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.8 | 8.8 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.8 | 7.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.7 | 2.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 2.1 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.7 | 7.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.7 | 3.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.7 | 2.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.6 | 2.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.6 | 7.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.6 | 7.9 | GO:0060013 | righting reflex(GO:0060013) |
| 0.5 | 12.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.5 | 3.5 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.5 | 1.9 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 | 2.3 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.4 | 1.3 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.4 | 4.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 1.3 | GO:0048369 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.4 | 14.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.4 | 3.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.4 | 1.8 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.4 | 2.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 3.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 1.1 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.3 | 1.7 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 | 5.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 4.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 2.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 1.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 0.3 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.3 | 1.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.3 | 8.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 5.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 2.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 4.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.3 | 0.8 | GO:0070602 | regulation of chondrocyte development(GO:0061181) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.3 | 7.5 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.2 | 0.7 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 2.2 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.2 | 0.7 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 5.7 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 2.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 14.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.2 | 2.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 2.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 4.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.2 | 0.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 5.2 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 14.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 1.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 2.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.6 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 1.6 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 4.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 0.4 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.2 | 5.9 | GO:0006260 | DNA replication(GO:0006260) |
| 0.2 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 24.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.6 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 3.0 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 3.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 2.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 2.4 | GO:0097150 | removal of superoxide radicals(GO:0019430) neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 2.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 5.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.8 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.7 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.6 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.2 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 2.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 2.6 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.1 | 3.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 3.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 0.9 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 2.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 3.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 2.8 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 4.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.8 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 8.4 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 1.2 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 2.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 3.0 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 48.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 4.8 | 24.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 3.8 | 57.7 | GO:0042555 | MCM complex(GO:0042555) |
| 3.3 | 23.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 2.4 | 12.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.8 | 16.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.8 | 5.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 1.6 | 4.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.5 | 5.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.5 | 4.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.3 | 9.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.2 | 15.0 | GO:0000800 | lateral element(GO:0000800) |
| 1.1 | 5.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.1 | 59.2 | GO:0005657 | replication fork(GO:0005657) |
| 1.1 | 1.1 | GO:0001740 | Barr body(GO:0001740) |
| 1.0 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 1.0 | 10.3 | GO:0000796 | condensin complex(GO:0000796) |
| 1.0 | 7.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.9 | 48.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.7 | 4.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 2.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.6 | 8.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.6 | 13.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.6 | 2.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.6 | 5.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 34.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 4.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 5.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.5 | 32.3 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.5 | 1.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 48.4 | GO:0005814 | centriole(GO:0005814) |
| 0.5 | 66.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.5 | 2.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 1.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 3.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 2.5 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 3.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 4.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 5.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 64.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 1.1 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 2.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 15.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 1.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 10.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 2.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 3.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 99.7 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 15.8 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 11.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 2.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 4.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 114.0 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 49.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 10.9 | 65.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 9.6 | 48.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 4.9 | 14.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 3.3 | 48.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 3.2 | 45.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 2.9 | 20.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 2.6 | 18.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 2.5 | 12.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 2.3 | 9.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 2.0 | 5.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.9 | 13.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.9 | 7.5 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 1.5 | 6.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 1.5 | 16.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.5 | 5.9 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 1.4 | 8.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 1.4 | 8.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.2 | 5.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 1.1 | 3.3 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 1.1 | 8.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.1 | 32.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.1 | 8.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.0 | 4.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.8 | 7.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.8 | 4.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.8 | 12.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.7 | 2.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.7 | 5.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.7 | 4.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.7 | 10.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.6 | 13.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 8.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.6 | 13.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 4.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.5 | 3.7 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.5 | 6.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 40.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.5 | 6.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 30.8 | GO:0000975 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.5 | 2.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.5 | 3.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.5 | 90.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.4 | 4.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.4 | 20.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.4 | 34.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 2.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 4.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.4 | 1.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.4 | 2.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 1.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.3 | 13.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 3.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 11.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 1.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.3 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 6.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 2.9 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 11.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 29.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 8.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 1.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.2 | 30.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 56.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 2.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 1.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 3.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 2.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.2 | 5.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 32.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 4.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.9 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 19.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 9.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 21.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 17.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 39.0 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 21.3 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 5.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 1.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.0 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 1.6 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 3.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 8.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 9.0 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 74.1 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 5.2 | 73.3 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 2.8 | 121.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 1.8 | 140.5 | PID_E2F_PATHWAY | E2F transcription factor network |
| 1.3 | 43.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.7 | 34.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.6 | 15.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.4 | 18.6 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.3 | 4.9 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 29.1 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 11.7 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.2 | 10.0 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 4.7 | PID_ATM_PATHWAY | ATM pathway |
| 0.2 | 3.7 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 2.0 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 8.3 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.2 | 8.4 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 7.3 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 1.6 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.2 | 2.0 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.7 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.2 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.3 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.8 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.1 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.2 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID_BMP_PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 175.3 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 8.4 | 92.1 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 4.5 | 63.6 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 3.5 | 31.9 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 2.5 | 41.7 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 2.1 | 8.4 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.8 | 12.9 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 1.7 | 23.4 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.5 | 17.9 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.1 | 30.4 | REACTOME_KINESINS | Genes involved in Kinesins |
| 1.0 | 5.8 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.6 | 33.0 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.5 | 2.5 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 3.0 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 11.0 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.4 | 5.8 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 7.3 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 8.4 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.3 | 5.1 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 57.8 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 5.2 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 10.8 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 5.0 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.3 | 9.1 | REACTOME_RNA_POL_I_TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.2 | 2.7 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 1.4 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 5.3 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.2 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 2.0 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 4.3 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.6 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 1.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 4.3 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 5.9 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 4.3 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.1 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.0 | REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 5.6 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.8 | REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 13.5 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.7 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 3.0 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.6 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |