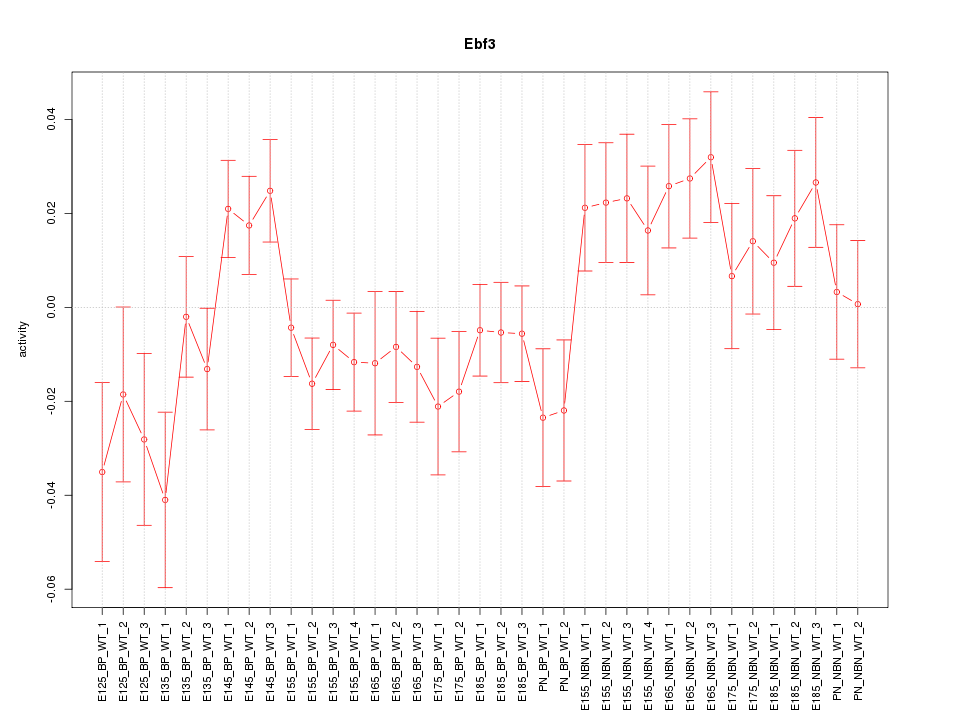
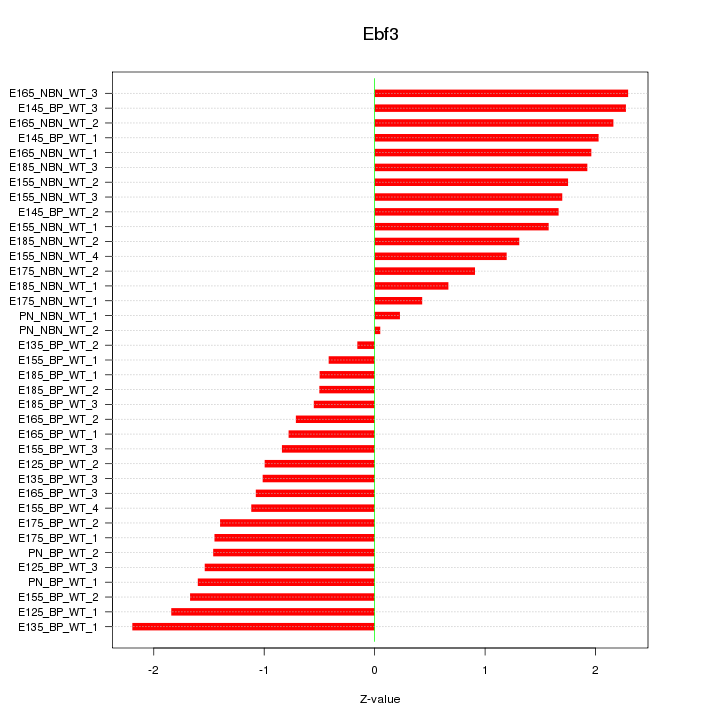
Motif ID: Ebf3
Z-value: 1.395
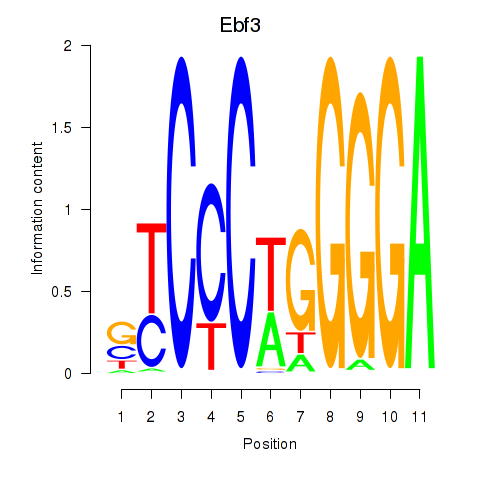
Transcription factors associated with Ebf3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ebf3 | ENSMUSG00000010476.7 | Ebf3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ebf3 | mm10_v2_chr7_-_137314394_137314445 | -0.65 | 1.2e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 21.9 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.2 | 6.7 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.0 | 6.0 | GO:0099548 | drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.2 | 7.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.2 | 19.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.0 | 3.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.0 | 4.8 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.9 | 3.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.9 | 5.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.9 | 4.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.8 | 5.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.8 | 1.6 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.8 | 3.8 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.7 | 8.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.7 | 4.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.7 | 2.6 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.7 | 4.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.7 | 2.6 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.6 | 2.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.6 | 1.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.5 | 1.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.5 | 2.1 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.5 | 1.0 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.5 | 2.5 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.5 | 4.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.5 | 3.8 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.5 | 0.9 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.5 | 2.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 1.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 1.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.4 | 4.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.4 | 3.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 1.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 2.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 1.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 1.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 2.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 4.8 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.3 | 1.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 1.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.3 | 0.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.3 | 4.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 1.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 2.0 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.2 | 1.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 0.7 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 5.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 4.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.2 | 1.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.0 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 | 1.0 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 1.0 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 0.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 2.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 3.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.7 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 4.8 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.2 | 12.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 2.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 0.5 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.2 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.2 | 4.0 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 | 3.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 2.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 | 1.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 4.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.9 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.1 | 1.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.1 | 2.4 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 1.9 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 2.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 3.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.3 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 1.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 12.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.2 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 6.0 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 2.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.6 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 0.4 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 1.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.3 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 1.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 1.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 1.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 2.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.8 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 4.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.5 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 1.1 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 2.2 | 21.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.5 | 4.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.1 | 4.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.1 | 5.3 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.8 | 2.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 2.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.7 | 8.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 6.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.5 | 6.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 2.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 1.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 3.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 4.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 3.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 2.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 3.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.0 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.2 | 3.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 15.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 6.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 6.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.1 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 19.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 10.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 3.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 2.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 7.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 12.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.9 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.0 | 6.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.3 | 4.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.2 | 5.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.1 | 7.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.1 | 5.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 1.0 | 3.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 4.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.8 | 8.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.8 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 4.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.7 | 2.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 5.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.6 | 4.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.5 | 3.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 19.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 1.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.5 | 1.9 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.4 | 1.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 1.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 5.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 3.8 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.3 | 1.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 12.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 0.8 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 1.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 1.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 2.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 2.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 1.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 1.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 4.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 2.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 3.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 3.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 4.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 5.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 3.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 3.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.0 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 4.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.8 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 10.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.9 | GO:0042277 | peptide binding(GO:0042277) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 21.9 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 5.8 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 2.1 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 10.4 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.2 | 25.5 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 5.5 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.0 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 4.0 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.5 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.1 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.9 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 10.0 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 3.4 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 0.7 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.3 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 1.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 4.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.7 | 15.1 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 11.5 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 8.8 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 3.1 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 1.2 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 8.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.2 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 6.0 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 7.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 10.1 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 6.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 2.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 5.8 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 3.8 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 3.8 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.3 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.1 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.6 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.5 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 4.2 | REACTOME_REGULATION_OF_INSULIN_SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 0.7 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.6 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.4 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.6 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.4 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.5 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.8 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME_CYCLIN_E_ASSOCIATED_EVENTS_DURING_G1_S_TRANSITION_ | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.3 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |