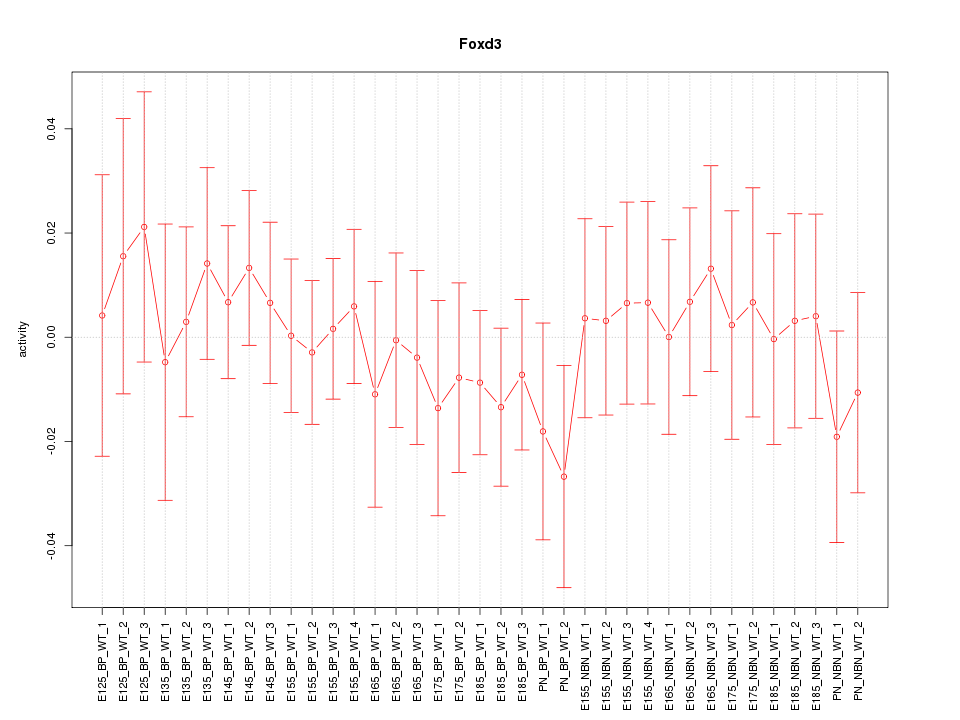
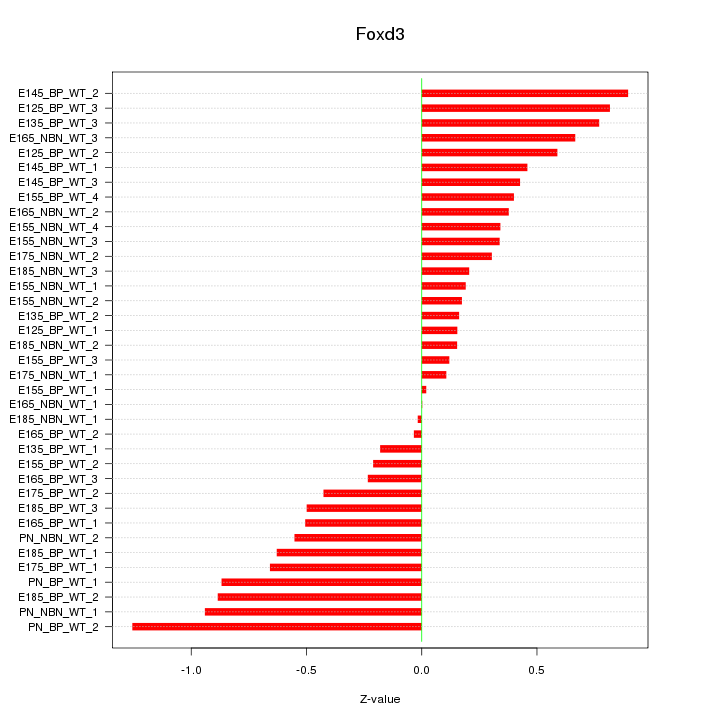
Motif ID: Foxd3
Z-value: 0.521
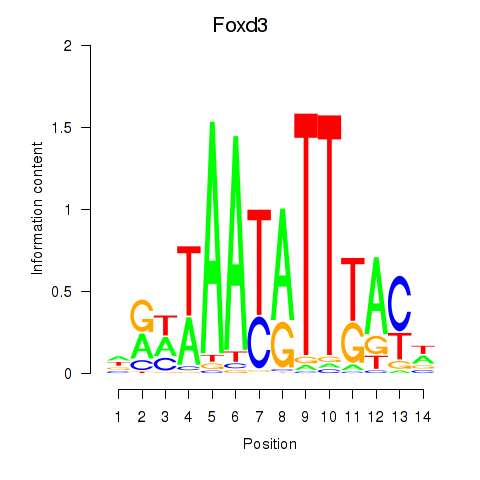
Transcription factors associated with Foxd3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxd3 | ENSMUSG00000067261.3 | Foxd3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.7 | 2.0 | GO:0071649 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 3.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 1.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.3 | 1.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 2.0 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.2 | 2.0 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 1.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.1 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 1.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0006547 | histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 1.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 2.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 1.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 5.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 2.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 1.4 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 2.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 5.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.7 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID_ALK2_PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.4 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.0 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.7 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.4 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 2.0 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |