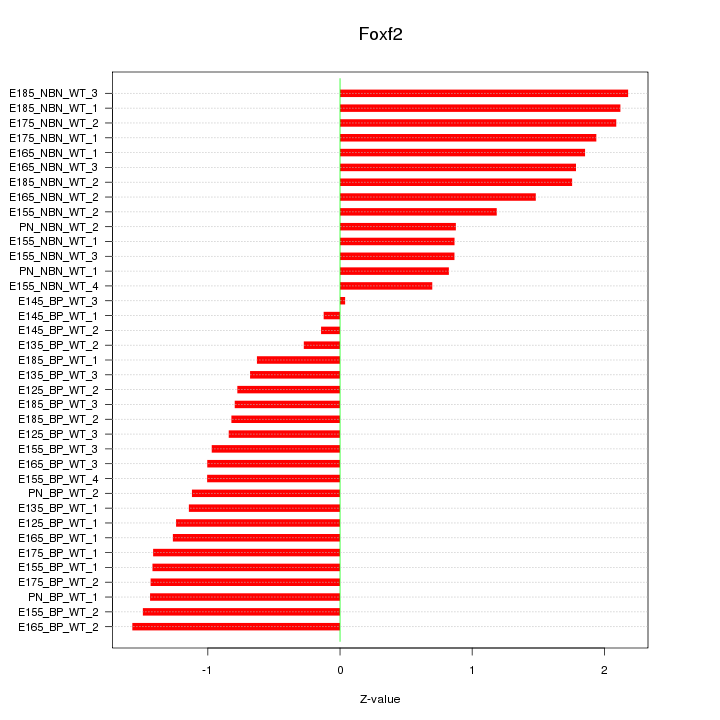
Motif ID: Foxf2
Z-value: 1.265
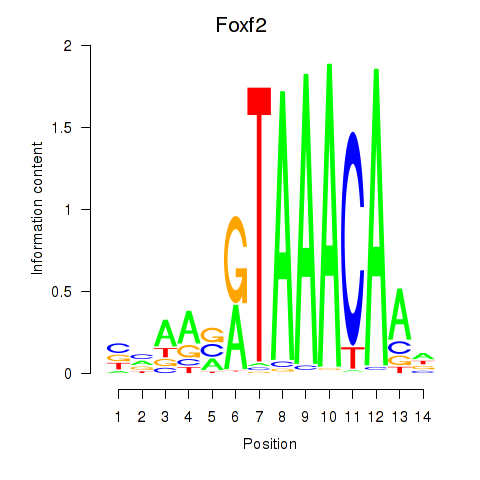
Transcription factors associated with Foxf2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxf2 | ENSMUSG00000038402.2 | Foxf2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf2 | mm10_v2_chr13_+_31625802_31625816 | 0.17 | 3.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.9 | 20.7 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 1.3 | 7.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.0 | 2.9 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 | 6.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 | 6.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.7 | 3.3 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.6 | 1.8 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.6 | 1.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.6 | 2.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 1.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 4.0 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.5 | 5.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.5 | 1.5 | GO:0043379 | rRNA export from nucleus(GO:0006407) memory T cell differentiation(GO:0043379) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.5 | 1.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.4 | 3.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.4 | 5.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 1.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.4 | 7.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 1.2 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 | 1.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.4 | 2.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 1.3 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.3 | 0.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.3 | 3.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.2 | 1.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 0.7 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.2 | 1.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 1.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 1.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 2.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 2.2 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 8.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.8 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 1.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 6.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 3.5 | GO:0051926 | dopamine receptor signaling pathway(GO:0007212) negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 0.7 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 | 1.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 2.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.5 | GO:0035826 | rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) |
| 0.1 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 1.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.8 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 1.4 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 15.6 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.7 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.6 | 21.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.5 | 1.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 1.3 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 3.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 1.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 2.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 3.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 17.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 6.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 7.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 6.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 24.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.6 | 1.8 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.6 | 3.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.6 | 2.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 20.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.5 | 1.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.4 | 2.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 2.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 1.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 3.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 4.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 6.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 5.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 7.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 22.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 2.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 12.0 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 20.7 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 3.5 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.2 | 6.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.0 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.5 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 5.0 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.3 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.6 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.9 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.3 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.0 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.1 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.3 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.0 | 0.6 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 1.9 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.0 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 4.2 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.5 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.5 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 9.0 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.3 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 3.5 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.8 | REACTOME_SIGNALLING_BY_NGF | Genes involved in Signalling by NGF |
| 0.1 | 0.6 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.1 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 4.3 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.8 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 3.3 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.5 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.5 | REACTOME_RNA_POL_I_RNA_POL_III_AND_MITOCHONDRIAL_TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.5 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.8 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |