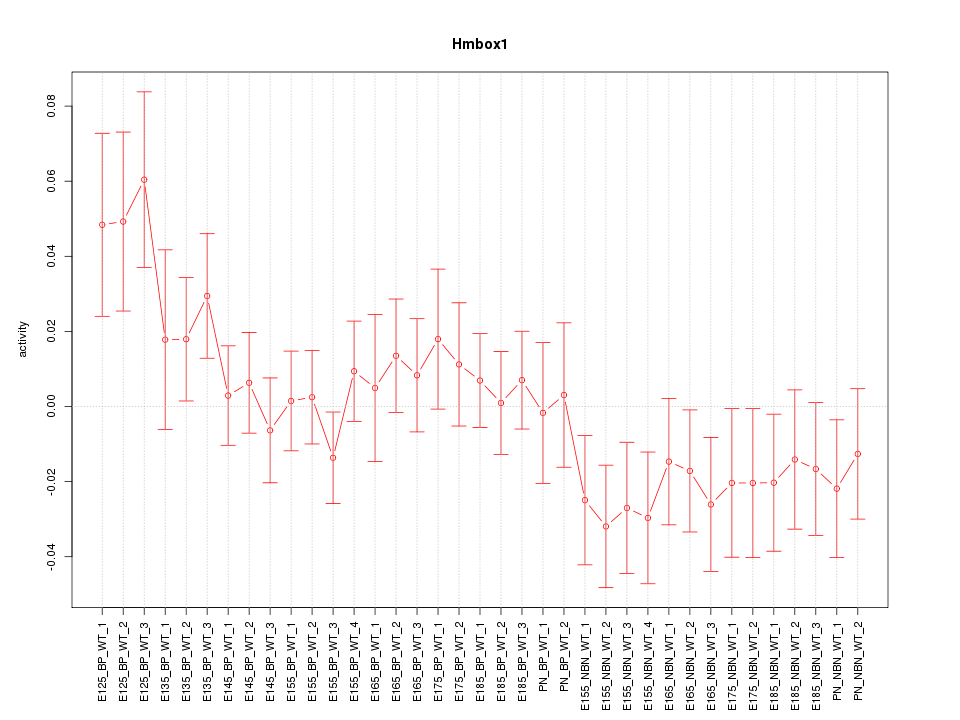
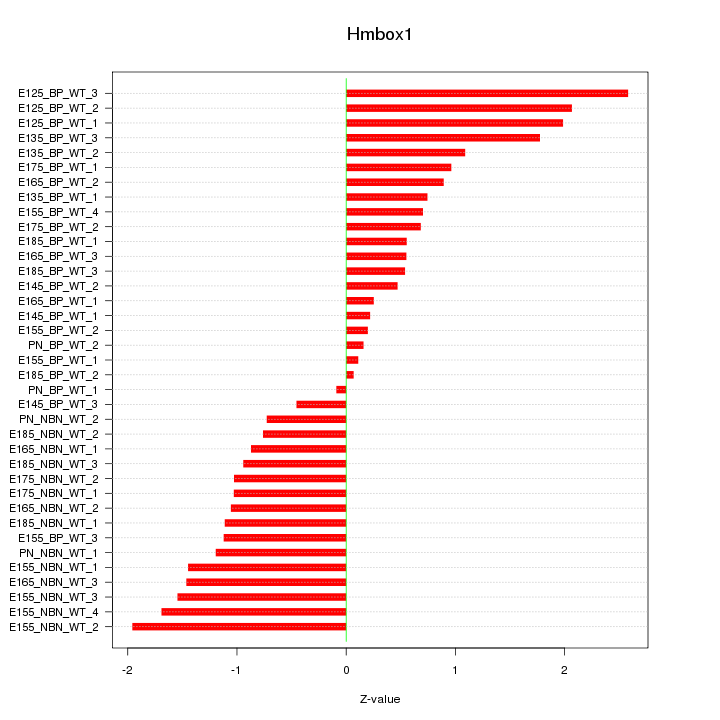
Motif ID: Hmbox1
Z-value: 1.130
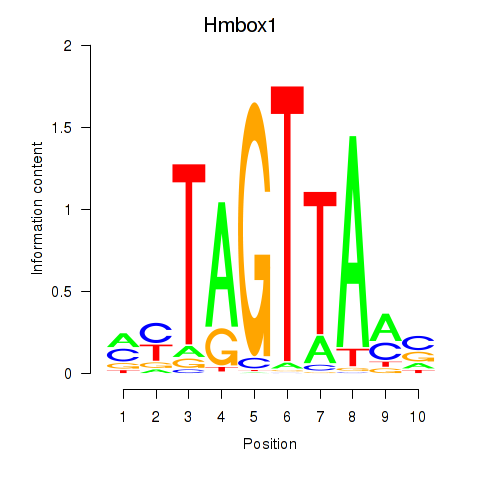
Transcription factors associated with Hmbox1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hmbox1 | ENSMUSG00000021972.8 | Hmbox1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmbox1 | mm10_v2_chr14_-_64949838_64949886 | 0.45 | 4.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 17.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.0 | 10.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.8 | 3.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.7 | 2.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.6 | 5.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 1.8 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 0.5 | 5.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.5 | 6.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 4.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 3.7 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 1.0 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.3 | 1.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.3 | 1.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 1.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 5.4 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.3 | 3.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.3 | 1.0 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.2 | 11.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 3.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.0 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 2.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.7 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.4 | GO:0099566 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.8 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 1.0 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.5 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.9 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 1.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 3.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.8 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.4 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 4.1 | GO:0022409 | positive regulation of cell-cell adhesion(GO:0022409) |
| 0.0 | 1.4 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.7 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 4.5 | GO:0009306 | protein secretion(GO:0009306) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.8 | 3.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 4.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.4 | 1.9 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 1.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 1.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 0.8 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 10.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 5.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 5.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 3.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 4.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 7.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 11.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 5.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 2.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.4 | 1.8 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 11.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.4 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 3.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 16.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 4.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 6.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.2 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 7.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 5.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.3 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 3.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 6.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 7.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 1.3 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.5 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 4.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 1.0 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 2.4 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 2.0 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.6 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.8 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.1 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.6 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.3 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.2 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 10.5 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.8 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |