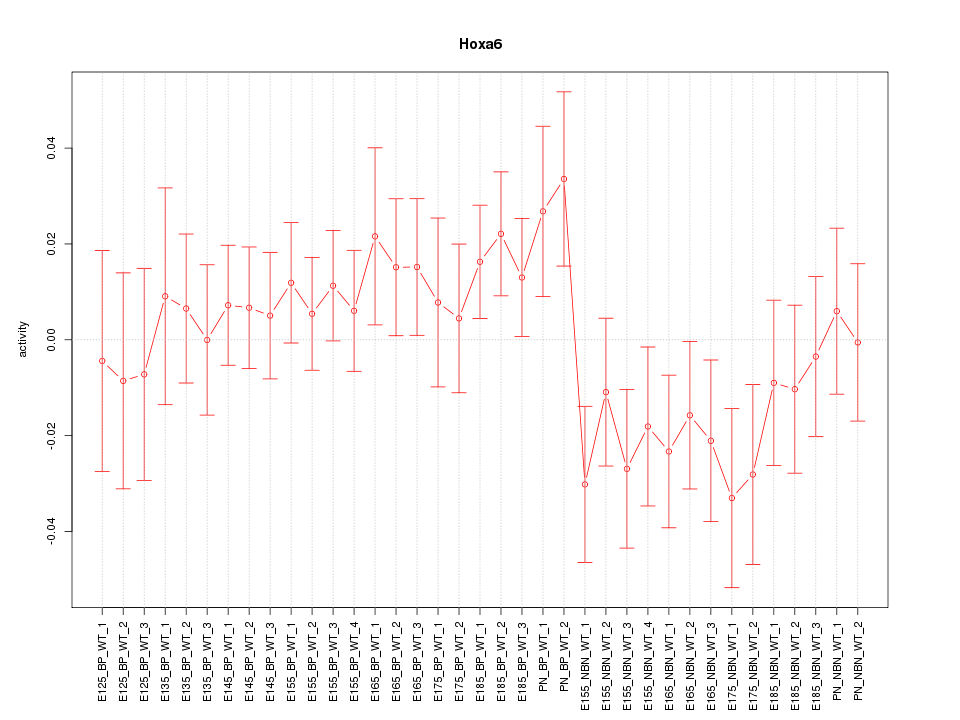
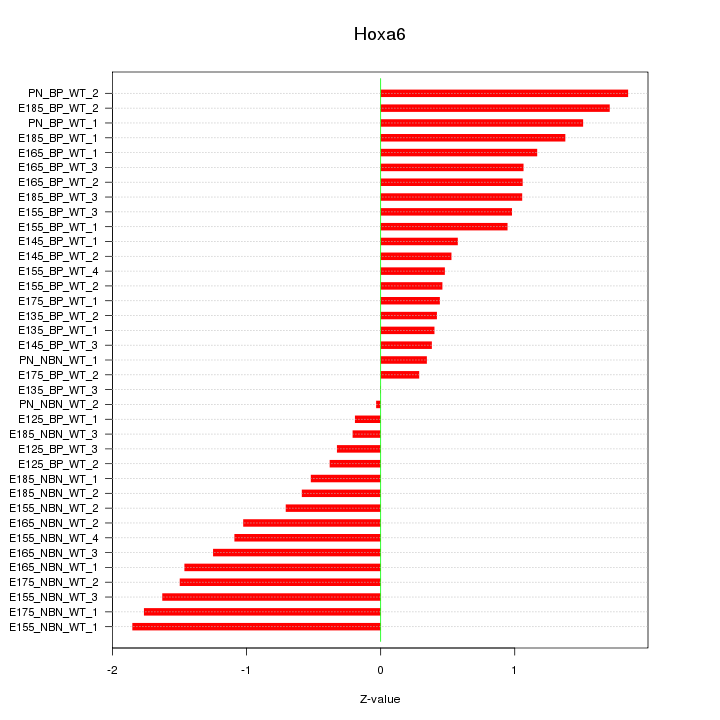
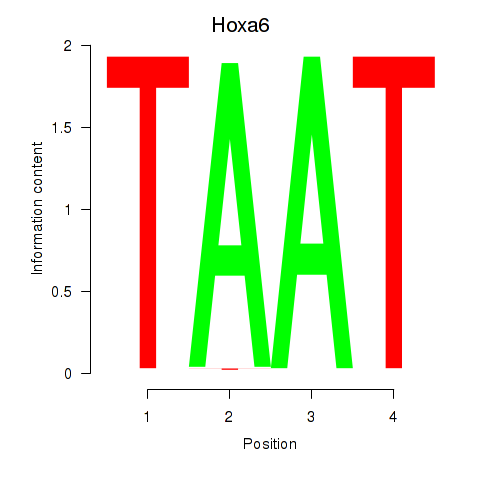
Motif ID: Hoxa6
Z-value: 1.012
Transcription factors associated with Hoxa6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa6 | ENSMUSG00000043219.8 | Hoxa6 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 2.1 | 14.9 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.9 | 5.8 | GO:0060023 | soft palate development(GO:0060023) |
| 1.9 | 3.8 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 1.7 | 3.4 | GO:0051884 | regulation of anagen(GO:0051884) |
| 1.6 | 6.5 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.1 | 6.6 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 1.1 | 3.2 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.9 | 13.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.9 | 3.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.8 | 10.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.8 | 2.4 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.8 | 3.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.8 | 5.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.8 | 5.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.8 | 9.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.7 | 8.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.7 | 4.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.7 | 7.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.6 | 25.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.5 | 1.1 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.5 | 1.5 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.5 | 6.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.5 | 4.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.5 | 0.9 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.4 | 1.3 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.4 | 1.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 6.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.3 | 3.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 0.5 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.2 | 0.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 2.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 1.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 4.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.2 | 2.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 5.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 0.2 | 9.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.2 | 0.7 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 3.2 | GO:1901629 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of presynaptic membrane organization(GO:1901629) |
| 0.2 | 0.8 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.7 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.6 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 8.3 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.5 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 2.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.3 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 2.5 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.3 | GO:1900141 | regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) |
| 0.1 | 0.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.8 | GO:0048733 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) sebaceous gland development(GO:0048733) |
| 0.1 | 7.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 3.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 7.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 3.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 1.2 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.8 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0045162 | protein localization to paranode region of axon(GO:0002175) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.4 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 1.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.5 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.0 | 8.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.9 | 8.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 2.9 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 7.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 6.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 2.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 18.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 26.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.3 | GO:0042567 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 8.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 2.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 8.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 5.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.3 | 3.8 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.9 | 12.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 6.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 58.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.3 | 21.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 5.5 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.3 | 3.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 8.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 7.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 4.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 4.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 4.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 13.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 3.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 3.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 2.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 36.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.1 | 1.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.0 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.8 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 12.2 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 7.1 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 3.4 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 9.1 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 14.6 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.2 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.4 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.7 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 2.4 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.2 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.1 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.4 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 14.6 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 3.3 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 8.3 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 0.9 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.4 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.4 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 7.4 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.7 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.6 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.7 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.8 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |