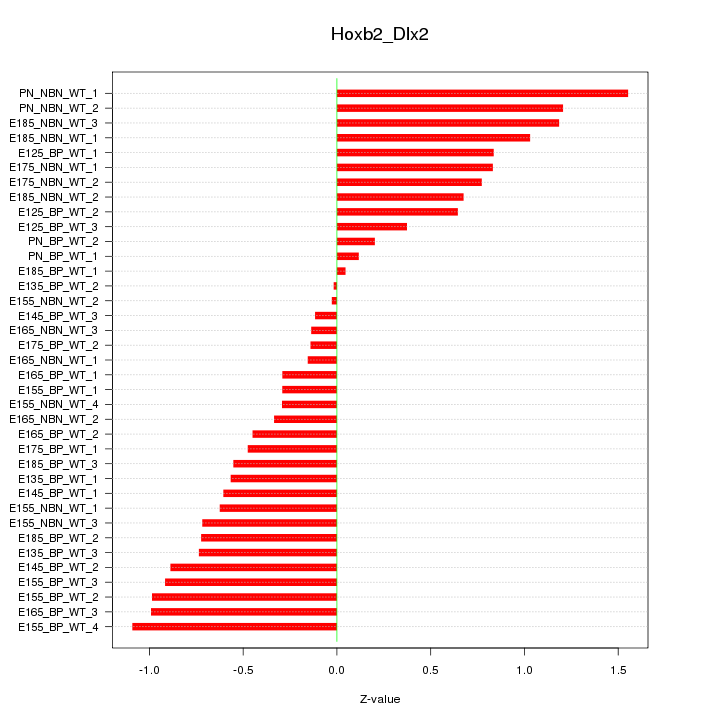
Motif ID: Hoxb2_Dlx2
Z-value: 0.698
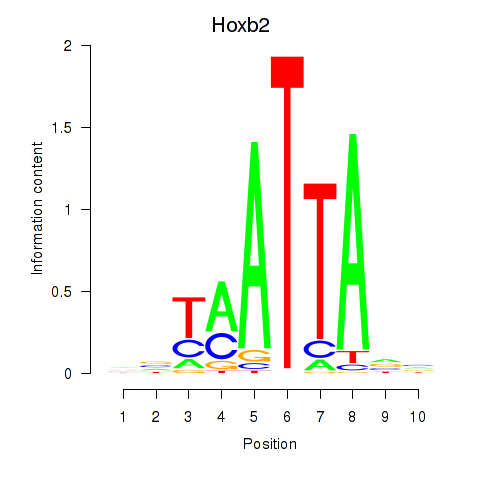
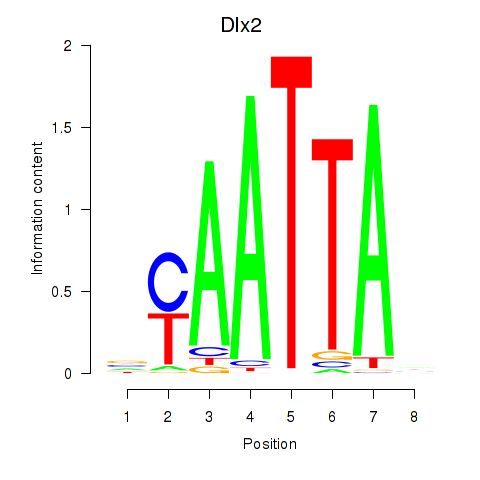
Transcription factors associated with Hoxb2_Dlx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dlx2 | ENSMUSG00000023391.7 | Dlx2 |
| Hoxb2 | ENSMUSG00000075588.5 | Hoxb2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx2 | mm10_v2_chr2_-_71546745_71546758 | 0.72 | 4.8e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 1.1 | 7.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.9 | 10.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.8 | 3.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.6 | 3.7 | GO:0045919 | complement activation, alternative pathway(GO:0006957) positive regulation of cytolysis(GO:0045919) |
| 0.4 | 2.6 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.4 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.4 | 1.4 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.3 | 1.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 0.9 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.3 | 1.2 | GO:0035937 | androgen catabolic process(GO:0006710) estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.3 | 0.9 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 0.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.3 | 0.8 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.3 | 1.1 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.3 | 1.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 0.7 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.9 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.2 | 0.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 0.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.3 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.2 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 1.4 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 2.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 0.5 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 | 1.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.4 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 2.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 1.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.4 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 2.0 | GO:0008210 | luteinization(GO:0001553) estrogen metabolic process(GO:0008210) |
| 0.1 | 0.5 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 2.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 1.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 2.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.7 | GO:0015838 | proline transport(GO:0015824) amino-acid betaine transport(GO:0015838) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 1.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 1.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 1.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.3 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 1.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 3.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.5 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.4 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 1.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.0 | GO:1904892 | JAK-STAT cascade(GO:0007259) regulation of JAK-STAT cascade(GO:0046425) STAT cascade(GO:0097696) regulation of STAT cascade(GO:1904892) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 2.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 0.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.3 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.7 | 7.4 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 2.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.3 | 1.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 2.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 0.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 0.9 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 1.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 1.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 1.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 9.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.0 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.3 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.7 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.3 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.8 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 1.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 3.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.4 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID_MYC_PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.7 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.6 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.0 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 9.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.0 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.3 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.2 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.2 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.4 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.0 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.8 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |