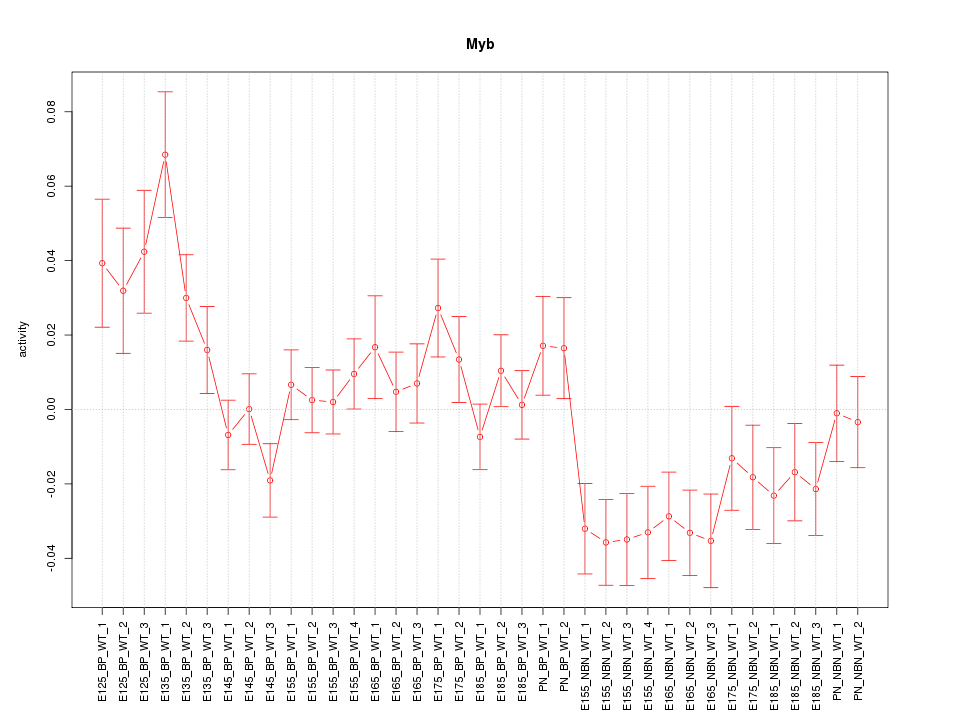
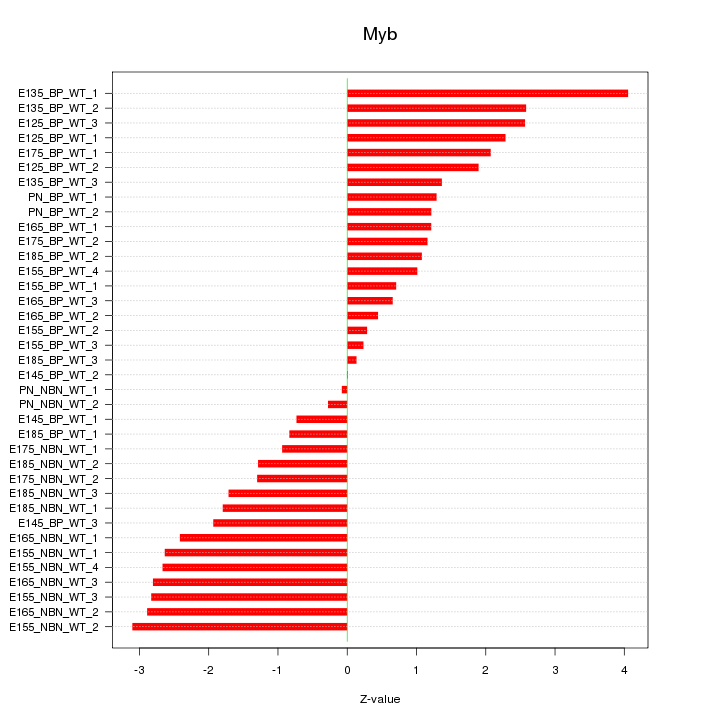
Motif ID: Myb
Z-value: 1.826
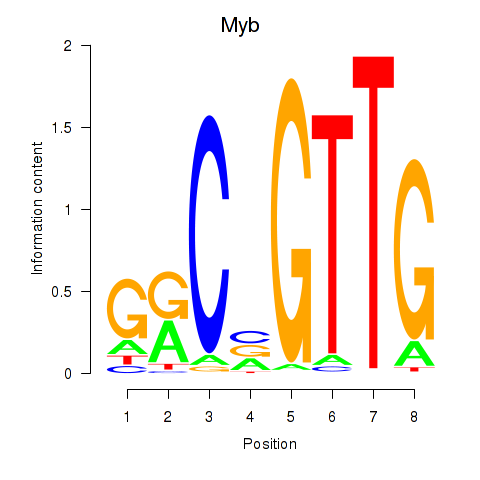
Transcription factors associated with Myb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Myb | ENSMUSG00000019982.8 | Myb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myb | mm10_v2_chr10_-_21160925_21160984 | 0.77 | 2.2e-08 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 3.8 | 11.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.9 | 17.7 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 2.2 | 6.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 2.0 | 35.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.9 | 13.4 | GO:0000279 | M phase(GO:0000279) |
| 1.5 | 6.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.5 | 7.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.2 | 12.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 1.1 | 7.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.0 | 16.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.0 | 16.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.9 | 7.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.9 | 7.9 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.8 | 2.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.8 | 0.8 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.8 | 1.5 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.7 | 2.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.7 | 2.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.7 | 3.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.7 | 2.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.6 | 3.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.6 | 2.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 | 1.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.5 | 1.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 2.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.5 | 1.5 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.5 | 3.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 3.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.5 | 2.0 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.5 | 3.4 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.4 | 2.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 2.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 2.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 5.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 5.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 3.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 4.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.4 | 2.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 3.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 6.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 2.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 1.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 3.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 9.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.3 | 2.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 1.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 0.8 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.3 | 3.9 | GO:0060430 | lung saccule development(GO:0060430) |
| 0.3 | 3.5 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.3 | 0.8 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 | 0.8 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.3 | 3.6 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.3 | 2.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 7.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 0.9 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.2 | 0.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 1.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 2.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 4.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 2.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.4 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 0.8 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 1.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 5.6 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 1.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.2 | 4.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 2.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.2 | 1.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.9 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 2.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 2.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.8 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 6.0 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 2.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 2.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.9 | GO:0043631 | mRNA polyadenylation(GO:0006378) mRNA 3'-end processing(GO:0031124) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.8 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.1 | 11.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 2.1 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 5.9 | GO:0007127 | meiosis I(GO:0007127) |
| 0.1 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.8 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 0.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.4 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 | 1.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 1.8 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 1.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.2 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 2.7 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.7 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 7.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 2.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 20.9 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.4 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 1.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 2.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.9 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 4.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.1 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.5 | GO:0043065 | positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.9 | 11.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 2.2 | 6.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.7 | 5.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.0 | 6.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 31.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.7 | 2.2 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.7 | 1.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.7 | 5.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.7 | 2.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.7 | 12.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.6 | 4.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 13.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 1.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 1.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 3.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 1.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 6.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 9.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 1.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 0.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.3 | 1.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 2.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.3 | 1.9 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.3 | 6.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 16.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 6.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 10.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 18.6 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 2.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 16.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 3.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 9.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 1.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 6.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 12.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 3.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 3.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 30.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 0.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 2.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 3.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.8 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 5.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 2.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 2.0 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.7 | 6.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.4 | 5.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.3 | 6.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.2 | 16.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 1.1 | 6.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.0 | 3.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.7 | 12.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.7 | 2.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.6 | 2.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.4 | 5.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 2.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.4 | 1.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 2.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 3.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 2.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 2.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 6.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 5.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 8.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 3.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 2.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 1.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 3.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 2.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 3.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 1.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 10.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 4.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 6.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 3.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.7 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 3.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 2.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 3.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 12.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 17.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 4.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 3.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 7.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 42.2 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.4 | 16.2 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.2 | 3.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 5.1 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 3.4 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.2 | 5.6 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.2 | 4.4 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 6.5 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.3 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.2 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.9 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.0 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.9 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 6.0 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 3.8 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.3 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.7 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.3 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.1 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 1.6 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 2.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.6 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID_ERBB1_RECEPTOR_PROXIMAL_PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.7 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.8 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.7 | 18.8 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.7 | 17.3 | REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.5 | 52.0 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 1.3 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 10.1 | REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 6.7 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 5.4 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.4 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 5.1 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 2.2 | REACTOME_COMPLEMENT_CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.6 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 2.0 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.0 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 6.6 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 3.3 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.6 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.9 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.8 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.9 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.7 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.6 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.6 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.7 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.5 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.9 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.8 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.5 | REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.4 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.7 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.6 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |