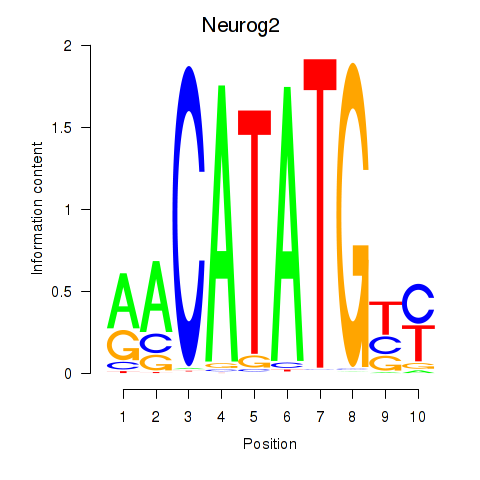
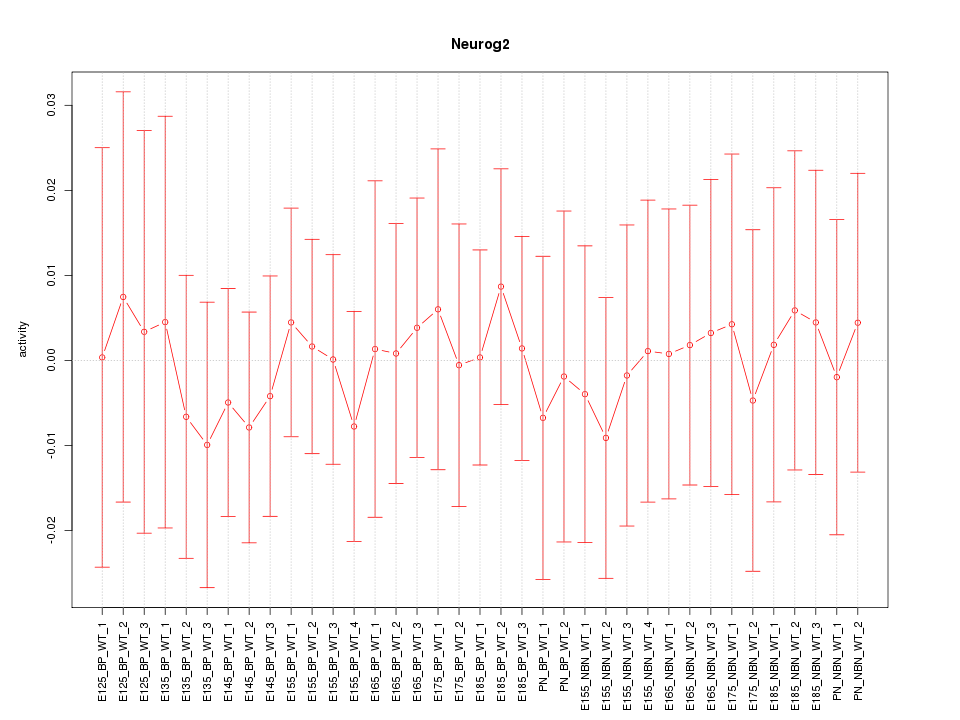
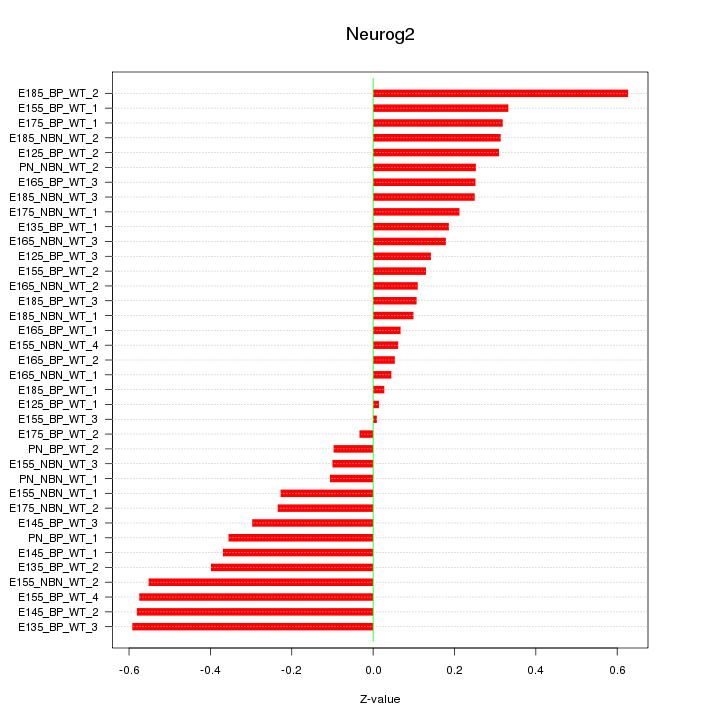
| 0.1 |
0.5 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) |
| 0.1 |
0.4 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.4 |
GO:0072236 |
DCT cell differentiation(GO:0072069) metanephric loop of Henle development(GO:0072236) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 |
0.2 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 |
0.4 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 |
0.2 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.2 |
GO:1903288 |
positive regulation of potassium ion import(GO:1903288) |
| 0.0 |
0.1 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 |
0.1 |
GO:2000097 |
regulation of smooth muscle cell-matrix adhesion(GO:2000097) negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 |
0.2 |
GO:0060857 |
establishment of glial blood-brain barrier(GO:0060857) |
| 0.0 |
0.2 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
0.6 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.2 |
GO:2000483 |
detection of bacterium(GO:0016045) response to bacterial lipoprotein(GO:0032493) detection of other organism(GO:0098543) negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
0.1 |
GO:0071544 |
diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.2 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.1 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 |
0.0 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 |
0.0 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.0 |
0.2 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 |
0.1 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.0 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |