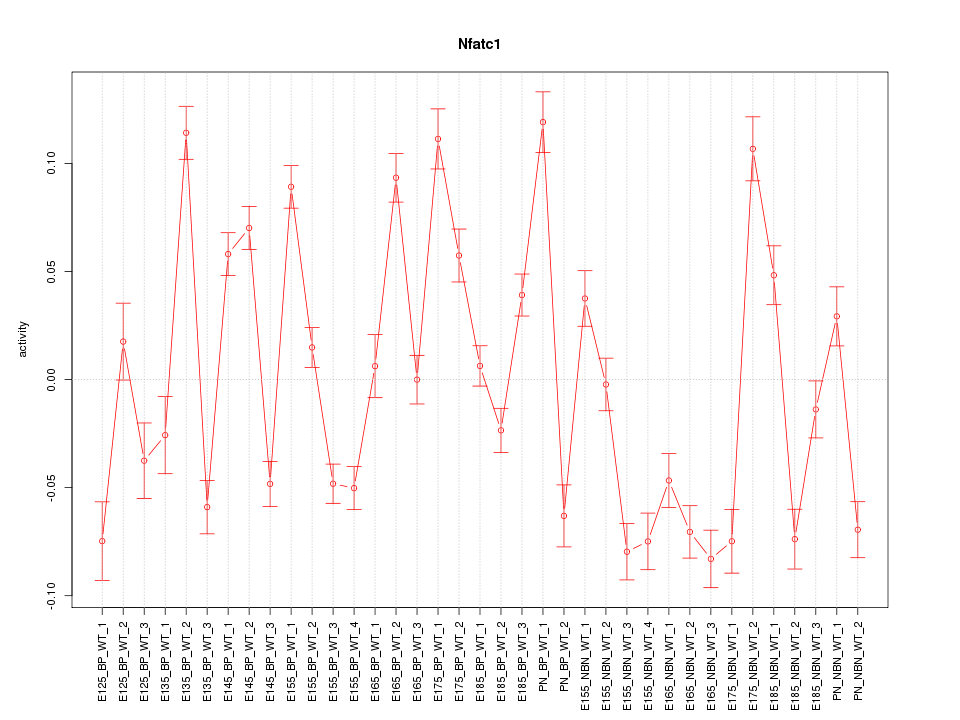
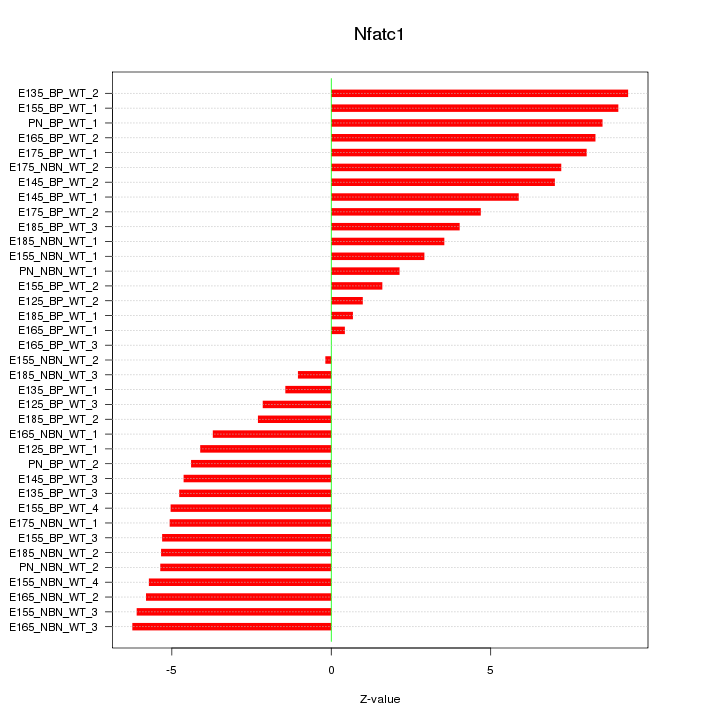
Motif ID: Nfatc1
Z-value: 5.102
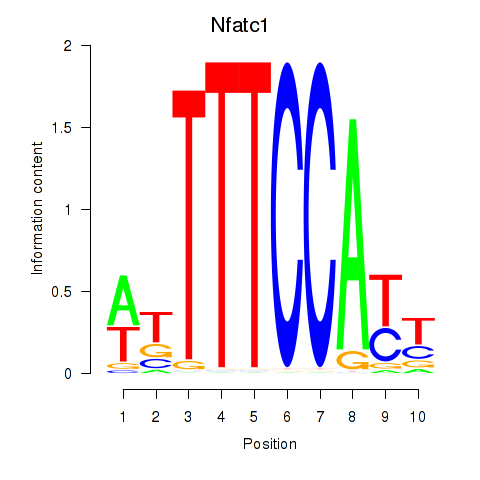
Transcription factors associated with Nfatc1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc1 | ENSMUSG00000033016.9 | Nfatc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm10_v2_chr18_-_80713062_80713080 | 0.19 | 2.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 3.2 | 12.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.0 | 6.0 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) negative regulation of bone development(GO:1903011) |
| 2.0 | 6.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.8 | 8.8 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 1.7 | 12.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.7 | 6.9 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.5 | 4.6 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.5 | 10.8 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.4 | 4.3 | GO:0099548 | drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.2 | 9.9 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.1 | 3.2 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 1.0 | 13.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.0 | 6.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 1.0 | 10.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.0 | 10.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.9 | 8.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.9 | 2.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.8 | 1.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.8 | 2.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) retinal rod cell differentiation(GO:0060221) intestinal epithelial structure maintenance(GO:0060729) renal vesicle induction(GO:0072034) |
| 0.7 | 2.9 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.7 | 4.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 2.8 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.7 | 7.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.7 | 5.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.6 | 2.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.6 | 1.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 2.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.5 | 2.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.5 | 13.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.5 | 1.4 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 1.4 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 1.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.4 | 1.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 2.0 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.4 | 2.6 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.4 | 1.1 | GO:0060558 | regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.3 | 3.8 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.3 | 1.9 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 2.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 0.9 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid metabolic process(GO:0046416) |
| 0.3 | 1.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 3.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 11.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 0.6 | GO:0090135 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.2 | 2.4 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 2.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 2.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 4.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.2 | 6.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 0.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.8 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 1.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.7 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.8 | GO:2000095 | common bile duct development(GO:0061009) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 1.5 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.8 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 1.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 1.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 4.0 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 4.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 4.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.1 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 1.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 1.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 864.2 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.2 | 4.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.9 | 2.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 2.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.4 | 4.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.4 | 8.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 8.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.4 | 4.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 3.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 4.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 3.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 3.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 9.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 3.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 6.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 3.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1069.6 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 1.4 | 4.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.2 | 4.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.2 | 4.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.0 | 3.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.0 | 6.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.0 | 6.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.0 | 3.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.0 | 19.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 6.0 | GO:0036122 | BMP binding(GO:0036122) |
| 0.5 | 4.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 2.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 1.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.4 | 2.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 2.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 12.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 3.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 1.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 2.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 4.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 6.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 4.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 3.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 9.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 3.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 4.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 3.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 10.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 997.4 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.8 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 23.2 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.3 | 10.1 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 12.5 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 2.6 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 2.3 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 6.0 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 17.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.4 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.8 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.2 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 9.0 | PID_P73PATHWAY | p73 transcription factor network |
| 0.1 | 3.4 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.6 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 1.4 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 2.8 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 0.8 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.7 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.7 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 4.0 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.7 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 23.4 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.5 | 4.6 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 2.5 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 7.0 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 2.3 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 1.6 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 9.9 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 1.4 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.3 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.3 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.8 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.4 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.4 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 9.6 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.6 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 9.5 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.1 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.9 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.7 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.1 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.5 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.0 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 3.1 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.3 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.2 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |