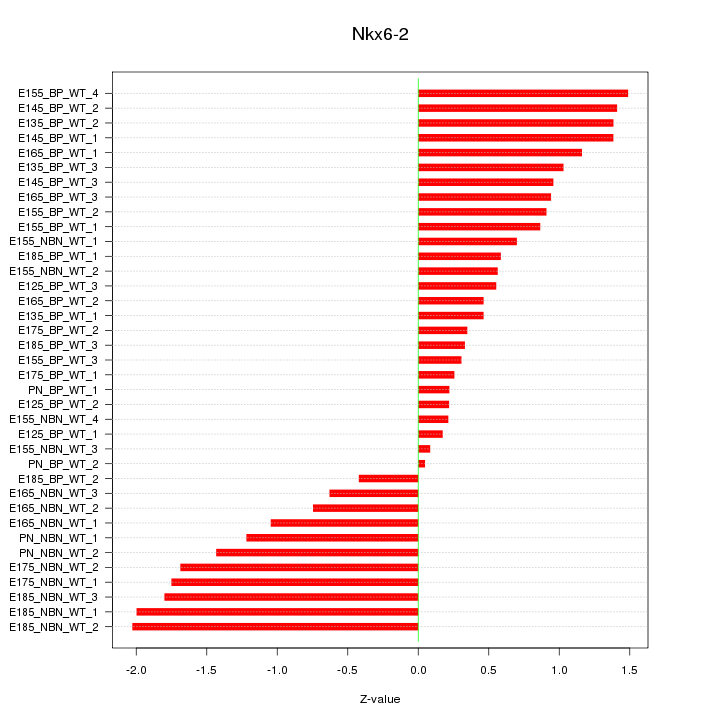
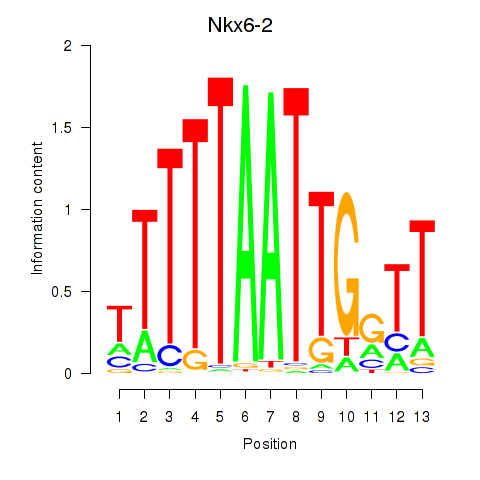
Motif ID: Nkx6-2
Z-value: 1.031
Transcription factors associated with Nkx6-2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nkx6-2 | ENSMUSG00000041309.11 | Nkx6-2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | mm10_v2_chr7_-_139582790_139582808 | -0.14 | 4.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0060023 | soft palate development(GO:0060023) |
| 1.4 | 11.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.1 | 3.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.9 | 4.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 2.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.6 | 2.8 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.5 | 2.0 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.4 | 1.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 1.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 2.0 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.3 | 1.7 | GO:0043654 | skeletal muscle satellite cell activation(GO:0014719) recognition of apoptotic cell(GO:0043654) |
| 0.3 | 2.7 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.3 | 1.6 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.3 | 4.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 0.5 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 1.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 1.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 1.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.6 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 1.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 8.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 0.9 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 1.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.4 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 0.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.6 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 3.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 3.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 3.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.4 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 2.4 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.1 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0006342 | chromatin silencing(GO:0006342) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 2.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 0.8 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 1.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 4.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.0 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 4.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 14.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 7.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 2.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.5 | 2.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 0.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 4.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 4.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 9.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 7.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 5.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 13.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.3 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 1.3 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.8 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.0 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 2.4 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 4.5 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 1.5 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 2.1 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 4.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 3.1 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.3 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.9 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 7.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.2 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.0 | REACTOME_CELL_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.6 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.0 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.4 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.4 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |