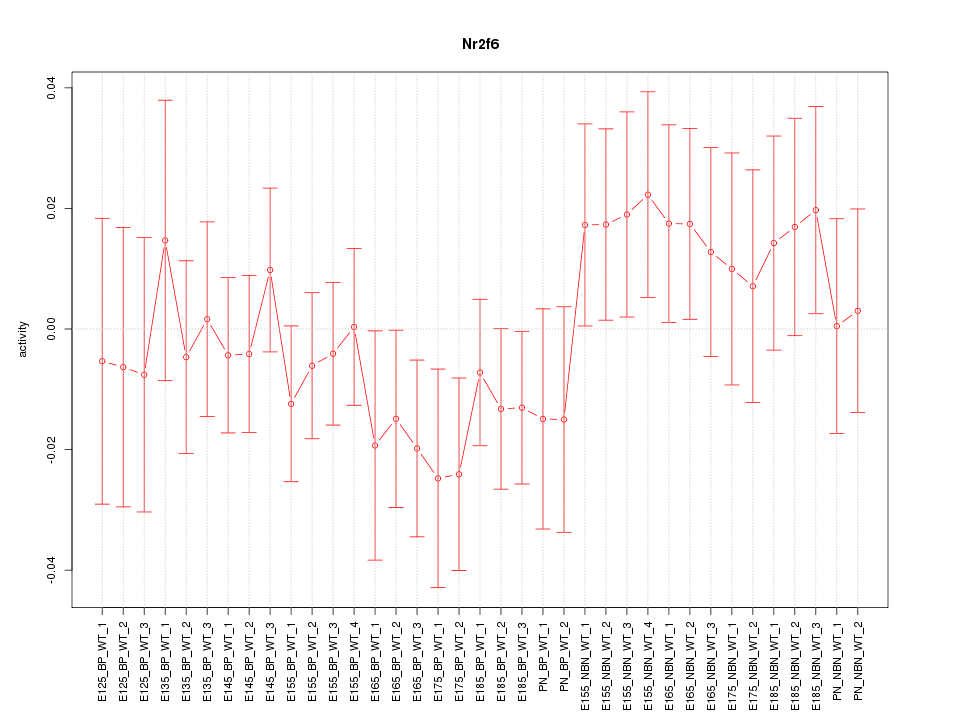
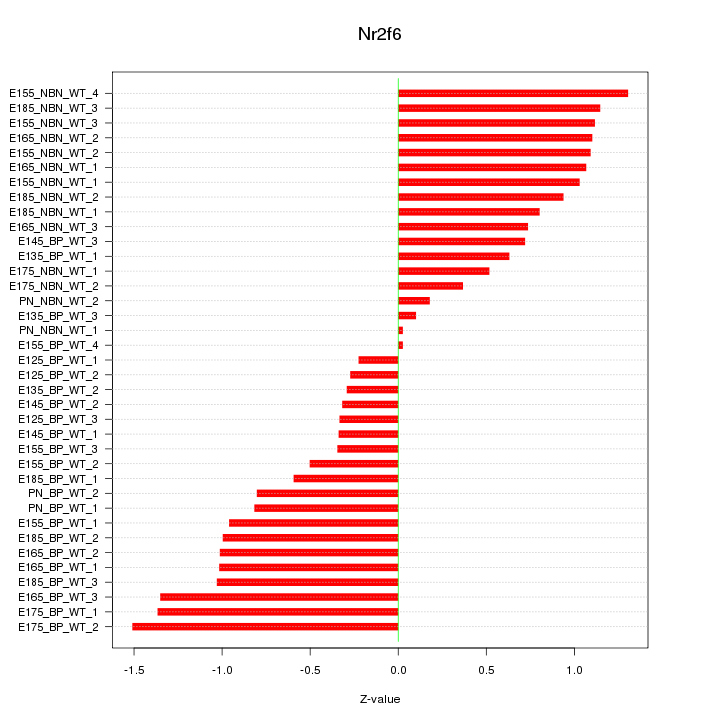
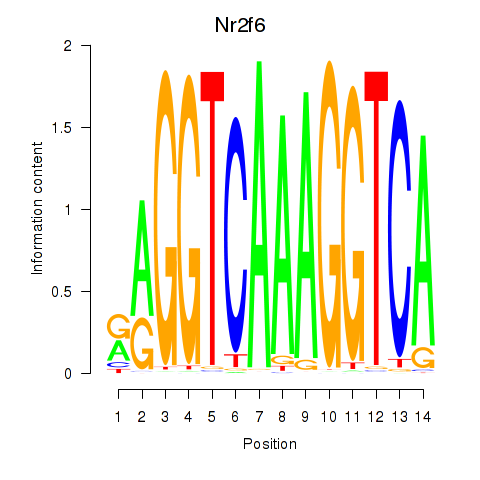
Motif ID: Nr2f6
Z-value: 0.837
Transcription factors associated with Nr2f6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr2f6 | ENSMUSG00000002393.8 | Nr2f6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f6 | mm10_v2_chr8_-_71381907_71381977 | 0.01 | 9.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.9 | 2.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 2.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 1.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.5 | 4.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 1.9 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.4 | 3.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 2.5 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 0.3 | 0.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 1.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.3 | 1.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 2.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 1.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 0.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.7 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.2 | 0.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 2.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.8 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 1.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.9 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.1 | 3.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.7 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 2.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 6.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.5 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.7 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.6 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 4.6 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 2.3 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 0.7 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.6 | 2.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 2.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.6 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 2.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 2.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 7.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.6 | 1.9 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.6 | 2.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.5 | 2.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 2.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 0.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 0.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 3.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 3.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 5.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 2.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.6 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 1.7 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.7 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.0 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 3.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.6 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.9 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.0 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.6 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 3.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.7 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.6 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.1 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 5.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.8 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |