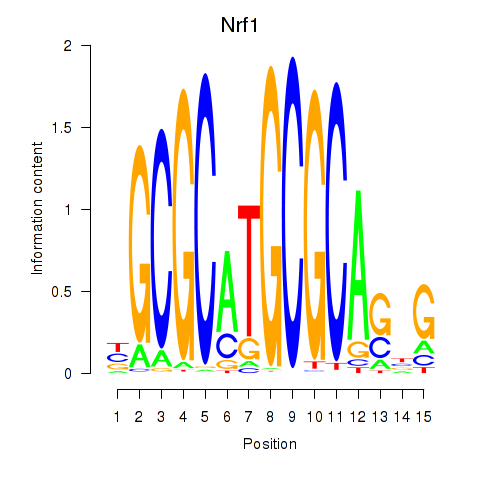
Motif ID: Nrf1
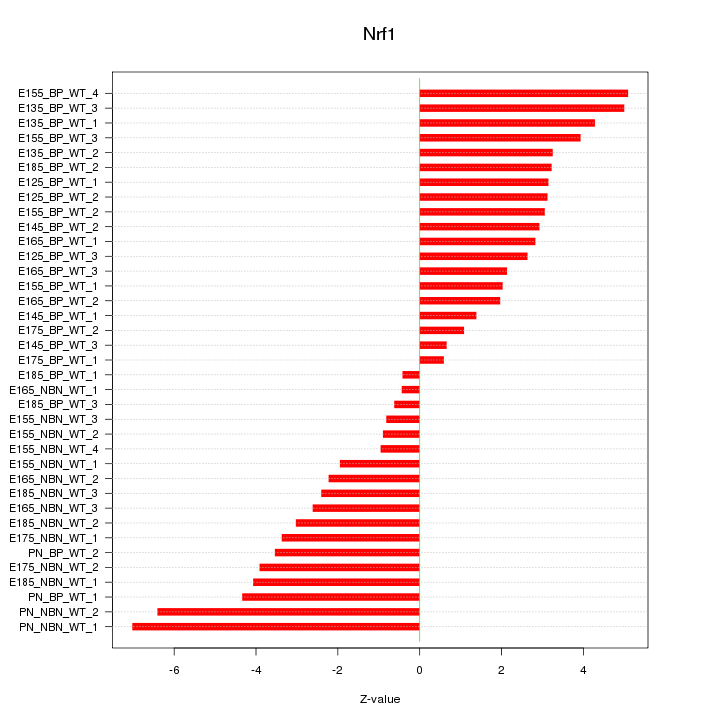
Z-value: 3.177
Transcription factors associated with Nrf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nrf1 | ENSMUSG00000058440.8 | Nrf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nrf1 | mm10_v2_chr6_+_30047979_30048049 | 0.74 | 1.3e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 3.8 | 3.8 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 3.7 | 14.7 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 3.3 | 13.0 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 2.6 | 23.6 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 2.4 | 7.2 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 2.0 | 10.2 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 2.0 | 6.0 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 2.0 | 2.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 2.0 | 7.9 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 1.9 | 15.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.8 | 5.5 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.8 | 5.5 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 1.8 | 5.4 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) |
| 1.8 | 5.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 1.8 | 3.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.7 | 7.0 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.7 | 10.5 | GO:0003383 | apical constriction(GO:0003383) |
| 1.7 | 8.6 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 1.6 | 6.5 | GO:2000256 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of male germ cell proliferation(GO:2000256) |
| 1.6 | 6.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.5 | 7.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.5 | 4.5 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 1.5 | 4.4 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 1.5 | 8.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.5 | 5.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 1.4 | 1.4 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 1.4 | 4.2 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.3 | 3.9 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.2 | 3.7 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 1.2 | 4.9 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 1.2 | 3.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 1.2 | 3.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.1 | 3.4 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 1.1 | 3.4 | GO:0003100 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of systemic arterial blood pressure by endothelin(GO:0003100) beta selection(GO:0043366) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 1.1 | 6.7 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 1.1 | 3.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.1 | 6.4 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 1.1 | 3.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.1 | 4.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 1.1 | 5.3 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 1.0 | 5.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 1.0 | 1.0 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 1.0 | 11.8 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 1.0 | 5.8 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.0 | 3.9 | GO:0032275 | luteinizing hormone secretion(GO:0032275) |
| 1.0 | 1.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.9 | 6.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.9 | 3.7 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.9 | 2.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.9 | 2.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.9 | 2.6 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.9 | 6.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.9 | 2.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.8 | 4.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.8 | 2.5 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.8 | 0.8 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.8 | 3.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 5.6 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.8 | 3.9 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.8 | 3.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.8 | 2.3 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.8 | 1.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.8 | 7.7 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.8 | 3.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 3.8 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.8 | 4.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.8 | 3.0 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.8 | 2.3 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.7 | 2.2 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.7 | 2.2 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.7 | 2.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.7 | 2.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.7 | 2.2 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.7 | 1.4 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.7 | 2.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.7 | 5.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.7 | 4.8 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.7 | 4.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.7 | 2.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.7 | 1.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.7 | 2.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.7 | 2.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.6 | 2.6 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.6 | 2.6 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.6 | 1.9 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.6 | 2.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.6 | 3.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.6 | 5.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.6 | 2.5 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.6 | 3.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.6 | 1.9 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.6 | 2.5 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.6 | 4.9 | GO:0015791 | polyol transport(GO:0015791) |
| 0.6 | 1.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.6 | 4.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.6 | 1.2 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.6 | 2.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.6 | 2.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.6 | 2.8 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.6 | 1.7 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.6 | 16.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.6 | 1.7 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.5 | 0.5 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.5 | 4.9 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.5 | 1.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.5 | 2.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.5 | 1.6 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.5 | 3.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 0.5 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.5 | 5.8 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.5 | 2.6 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.5 | 0.5 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.5 | 2.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.5 | 2.1 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.5 | 1.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.5 | 2.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.5 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.5 | 2.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.5 | 2.0 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.5 | 2.5 | GO:0019230 | proprioception(GO:0019230) |
| 0.5 | 6.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.5 | 2.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.5 | 2.0 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.5 | 0.5 | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.5 | 1.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 1.4 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.5 | 2.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.5 | 2.9 | GO:0042148 | strand invasion(GO:0042148) |
| 0.5 | 3.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.5 | 0.9 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.5 | 0.9 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.4 | 0.9 | GO:0097212 | late endosomal microautophagy(GO:0061738) lysosomal membrane organization(GO:0097212) |
| 0.4 | 5.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.4 | 3.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.3 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.4 | 2.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 2.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 1.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.4 | 3.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 1.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 0.8 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.4 | 2.5 | GO:0070922 | small RNA loading onto RISC(GO:0070922) |
| 0.4 | 1.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.4 | 1.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.4 | 6.8 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.4 | 6.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.4 | 1.6 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 0.8 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.4 | 1.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 1.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.4 | 1.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.4 | 5.0 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.4 | 2.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.4 | 12.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 5.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 1.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.4 | 8.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.4 | 1.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.4 | 1.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.4 | 2.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 7.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 0.4 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.4 | 2.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.4 | 1.4 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.4 | 2.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.4 | 2.2 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.4 | 5.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.4 | 1.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 1.4 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.4 | 1.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 1.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 0.7 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.3 | 1.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 1.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.3 | 1.0 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.3 | 2.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.3 | 1.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.3 | 4.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.3 | 0.7 | GO:0061525 | hindgut development(GO:0061525) |
| 0.3 | 5.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 1.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.3 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.3 | 2.6 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.3 | 2.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 1.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 1.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 3.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 0.9 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.3 | 3.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 0.9 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 | 1.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 1.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 1.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 2.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 1.2 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.3 | 2.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 1.5 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.3 | 3.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 0.9 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.3 | 0.9 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.3 | 1.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 6.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.3 | 2.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 1.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 0.8 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 1.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.3 | 3.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 1.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.3 | 0.6 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.3 | 1.6 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.3 | 1.9 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.3 | 1.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 4.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 3.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 2.1 | GO:0019359 | NAD biosynthetic process(GO:0009435) nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.3 | 2.9 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 2.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 4.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.7 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.7 | GO:0006272 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.2 | 0.5 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.2 | 4.3 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.2 | 1.7 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.2 | 1.7 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 2.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 5.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 3.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 1.4 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.2 | 0.9 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 1.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 1.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 4.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 6.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 0.9 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.2 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 1.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 1.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 0.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 0.9 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 1.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 0.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 1.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 2.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.8 | GO:0072362 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 | 0.8 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.6 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.2 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 3.5 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.2 | 1.6 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 0.6 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 0.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 4.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 0.6 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 1.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 2.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 0.8 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.2 | 1.3 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.2 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 1.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 4.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.2 | 1.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.2 | 4.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.5 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 0.3 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.2 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 0.7 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 1.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 0.2 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.2 | 1.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 1.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 2.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 0.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.5 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.2 | 0.3 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.2 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 1.8 | GO:0060538 | skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.2 | 0.8 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 0.5 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 1.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 2.1 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 8.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 0.9 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 2.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.9 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 1.6 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.3 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 6.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.9 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 2.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 2.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 3.8 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.1 | 0.4 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 1.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.0 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 0.9 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 2.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 6.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 5.5 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 5.1 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.1 | 0.4 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 1.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.4 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 3.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.9 | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.1 | 1.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.1 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 1.6 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.1 | 2.7 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 1.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.6 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 1.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.9 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 1.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 3.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 2.1 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 4.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.7 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 1.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.4 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.5 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 2.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 1.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 5.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.9 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 1.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.6 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 4.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.7 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 1.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 1.0 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 1.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.9 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.7 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.9 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 1.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.7 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.4 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.1 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.8 | GO:0003170 | heart valve development(GO:0003170) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 1.4 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 0.8 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 2.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 0.2 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.1 | 0.9 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 1.9 | GO:0098868 | endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.1 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.5 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 1.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 0.6 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 1.0 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 1.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 5.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.7 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.1 | 0.7 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.1 | GO:1902415 | regulation of mRNA binding(GO:1902415) positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.9 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 1.5 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.1 | 1.9 | GO:1904591 | positive regulation of protein import(GO:1904591) |
| 0.1 | 0.9 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.6 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.1 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 5.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 0.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.6 | GO:1904152 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.8 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.3 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 2.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.4 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 1.3 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 1.0 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 1.4 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 2.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.6 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.1 | 2.5 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 5.6 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 2.1 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.0 | GO:0060042 | retina morphogenesis in camera-type eye(GO:0060042) |
| 0.1 | 0.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.4 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.1 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.7 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.1 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.1 | 1.2 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 1.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.1 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.3 | GO:0051893 | regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.5 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 1.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.0 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.5 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.0 | 0.3 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 2.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.6 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 | 0.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.3 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 9.1 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0010042 | response to manganese ion(GO:0010042) response to cobalt ion(GO:0032025) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.8 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.7 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0002757 | immune response-activating signal transduction(GO:0002757) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.6 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.5 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 1.5 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 3.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.5 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.5 | GO:0071103 | DNA conformation change(GO:0071103) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.7 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.3 | GO:0052548 | regulation of endopeptidase activity(GO:0052548) |
| 0.0 | 0.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.7 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.3 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.0 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.0 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.3 | GO:0000939 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 3.0 | 8.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 2.8 | 8.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 2.8 | 8.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 2.2 | 4.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 1.5 | 10.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 1.4 | 10.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.3 | 6.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.3 | 17.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.3 | 7.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.2 | 9.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.1 | 5.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.1 | 7.6 | GO:0000796 | condensin complex(GO:0000796) |
| 1.0 | 6.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 5.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.9 | 3.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.8 | 2.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.8 | 4.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.7 | 1.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.7 | 2.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.7 | 3.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.7 | 4.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.7 | 7.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.7 | 2.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.6 | 1.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.6 | 3.1 | GO:0001652 | granular component(GO:0001652) |
| 0.6 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.6 | 2.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.6 | 0.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.6 | 7.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.6 | 1.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.5 | 2.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 2.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.5 | 3.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 2.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.5 | 3.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.5 | 2.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 2.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.5 | 6.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.5 | 8.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 10.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.5 | 9.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 3.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 3.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 2.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 6.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 2.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 6.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 1.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.4 | 0.8 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.4 | 1.7 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.4 | 1.7 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.4 | 13.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.4 | 5.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 3.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 2.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 2.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 1.4 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.4 | 1.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 7.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.4 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 3.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.3 | 11.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 0.3 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.3 | 3.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 1.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 7.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 40.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.3 | 1.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 2.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.3 | 2.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 2.3 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.3 | 2.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 1.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.3 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 0.8 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.3 | 4.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.0 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 12.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 4.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 1.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 2.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 2.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 6.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 9.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 2.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 18.1 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 1.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 2.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 16.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 1.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.4 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.2 | 1.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 2.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 2.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 6.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.2 | 1.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 1.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 0.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 0.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 1.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.4 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 7.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 7.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 3.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.9 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 8.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 4.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 6.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 8.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 13.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 1.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 2.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 12.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 2.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 11.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 3.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 15.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 7.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 7.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 1.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 1.7 | 6.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.7 | 5.0 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 1.7 | 8.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.6 | 4.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.5 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.3 | 6.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.3 | 7.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.2 | 4.7 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.1 | 3.3 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.1 | 5.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.1 | 13.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.0 | 3.9 | GO:0031493 | nucleosomal histone binding(GO:0031493) hemi-methylated DNA-binding(GO:0044729) |
| 0.9 | 3.7 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.9 | 3.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.9 | 2.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 2.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.9 | 13.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.9 | 6.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.8 | 3.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.8 | 5.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.7 | 2.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.7 | 2.9 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.7 | 2.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.7 | 6.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.7 | 2.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.7 | 2.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.7 | 0.7 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.7 | 3.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.7 | 4.8 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.7 | 2.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.7 | 3.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.6 | 2.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.6 | 2.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.6 | 3.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 4.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 3.7 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.6 | 1.8 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.6 | 5.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.6 | 2.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 1.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.6 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 5.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.6 | 3.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.6 | 2.2 | GO:0016531 | copper chaperone activity(GO:0016531) cuprous ion binding(GO:1903136) |
| 0.5 | 3.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.5 | 2.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.5 | 9.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 3.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.5 | 3.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.5 | 2.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.5 | 2.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 2.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.5 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.5 | 0.5 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.5 | 1.4 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.5 | 1.9 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.5 | 3.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 3.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 3.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 2.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 1.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 1.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 15.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 3.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 1.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 3.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 3.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 2.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 2.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 4.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.6 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.4 | 1.6 | GO:0004844 | mismatch base pair DNA N-glycosylase activity(GO:0000700) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.4 | 2.0 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.4 | 6.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 4.3 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.4 | 0.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.4 | 2.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 1.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 1.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.4 | 1.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.4 | 1.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 1.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.4 | 1.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 17.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 1.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 5.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 5.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 6.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 1.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 2.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.4 | 2.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.4 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.3 | 3.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 2.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 1.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.3 | 4.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 3.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 10.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 0.9 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.3 | 0.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 5.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 15.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 1.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.3 | 2.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 0.9 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.3 | 0.9 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 5.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.9 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.3 | 1.5 | GO:0010340 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 0.9 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.3 | 3.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 1.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 3.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 2.0 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 0.8 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.3 | 2.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 1.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 3.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 6.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 2.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 0.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 1.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 1.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 4.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 0.8 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.3 | 1.8 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.3 | 0.8 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.3 | 4.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 3.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 11.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 4.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 5.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 0.7 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 0.7 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 8.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.9 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 2.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 4.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 1.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 1.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.2 | 1.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.2 | 1.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 2.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 3.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 6.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 11.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 1.6 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 0.6 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.9 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 1.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.8 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 2.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 2.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.2 | 12.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 0.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 0.9 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 14.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 2.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 5.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 2.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.8 | GO:0047134 | thioredoxin-disulfide reductase activity(GO:0004791) protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 4.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 0.6 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.2 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 3.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 4.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 4.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 0.9 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) activin-activated receptor activity(GO:0017002) |
| 0.1 | 2.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 7.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 0.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 8.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 6.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.3 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.1 | 0.3 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 9.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 5.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 3.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.7 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 1.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 12.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 4.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 3.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 4.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 2.1 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 4.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 3.4 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.7 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.2 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 34.7 | GO:0001012 | RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.1 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.1 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 1.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 1.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.0 | GO:0008186 | RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 3.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 3.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 4.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 3.4 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 5.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 4.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 40.4 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.7 | 9.9 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.6 | 8.6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 2.9 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 1.6 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.5 | 4.1 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 5.5 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.5 | 22.1 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.4 | 11.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 5.6 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 20.5 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.3 | 7.2 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.3 | 7.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.3 | 6.8 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 23.5 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.2 | 2.9 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 1.1 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 2.1 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 2.5 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 1.4 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 5.0 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.2 | 2.9 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 3.7 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.2 | 1.8 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.6 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.8 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 8.8 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.5 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.8 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.1 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 10.7 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.8 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.4 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.0 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.0 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 7.2 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 6.0 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 10.5 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 5.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 4.4 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.3 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 0.9 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.1 | 3.9 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.8 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 1.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 1.7 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 1.3 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.0 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 2.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.3 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 2.2 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.9 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.6 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.6 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 1.3 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.1 | 3.2 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.9 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.2 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.8 | 16.3 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.8 | 7.8 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.8 | 15.9 | REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.6 | 7.1 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.6 | 4.7 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 18.1 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.5 | 53.3 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.5 | 6.0 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.5 | 8.1 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.5 | 5.7 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.5 | 4.2 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 3.5 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 7.0 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 0.4 | REACTOME_P53_DEPENDENT_G1_DNA_DAMAGE_RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.4 | 1.1 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 2.0 | REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.3 | 14.2 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 11.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 0.3 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 4.4 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 4.0 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.3 | 1.2 | REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.3 | 2.1 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 4.1 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 2.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 2.2 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 2.5 | REACTOME_SCFSKP2_MEDIATED_DEGRADATION_OF_P27_P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.3 | 1.6 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.3 | 6.5 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.3 | 8.0 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.3 | 2.0 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 2.0 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 2.2 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 5.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.2 | 1.7 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.3 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 1.9 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.3 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 5.7 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 11.2 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 0.8 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.2 | 4.6 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.2 | 1.3 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 7.6 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 5.5 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 2.4 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.2 | 5.3 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 1.0 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 4.1 | REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.2 | 4.8 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 30.9 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 2.0 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.2 | 6.3 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 5.1 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 2.1 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 6.1 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 2.0 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 5.1 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 3.5 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 7.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 4.0 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.3 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.5 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.9 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.1 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 0.4 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.8 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.8 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.4 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.9 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.3 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.6 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 5.2 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.9 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.7 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.8 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.2 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 4.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.6 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 0.4 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 3.9 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.0 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_ACTIVATES_SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 0.2 | REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.3 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 4.1 | REACTOME_PI_METABOLISM | Genes involved in PI Metabolism |
| 0.1 | 1.7 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.1 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 3.9 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 5.2 | REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 0.6 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.9 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.8 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.1 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.1 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.5 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.3 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.6 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME_PROCESSING_OF_CAPPED_INTRONLESS_PRE_MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.3 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME_HIV_LIFE_CYCLE | Genes involved in HIV Life Cycle |
| 0.0 | 0.2 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.3 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |