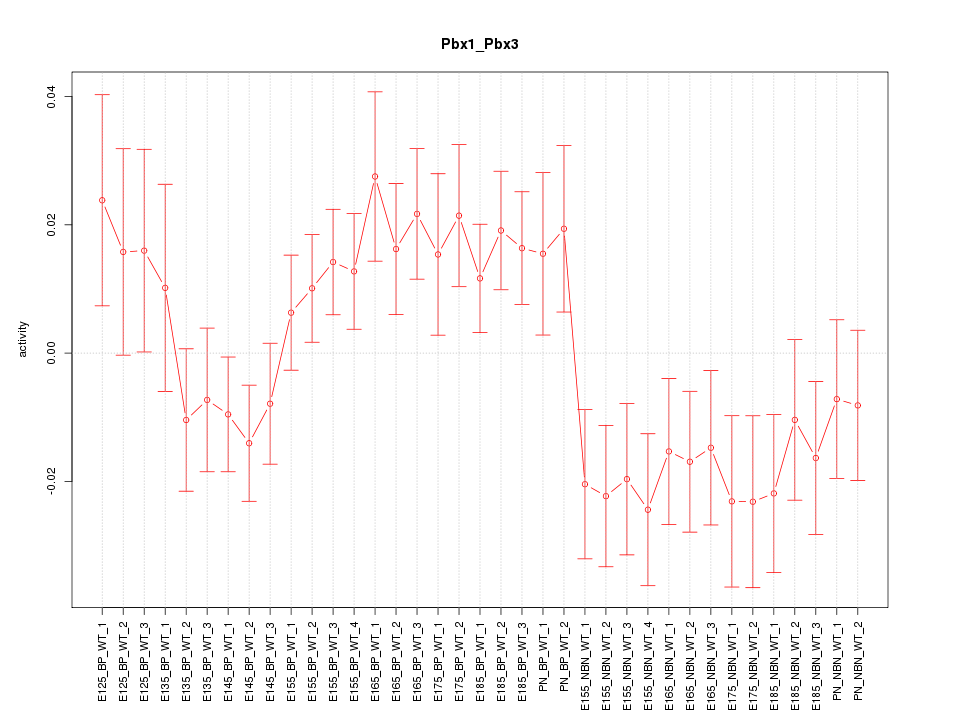
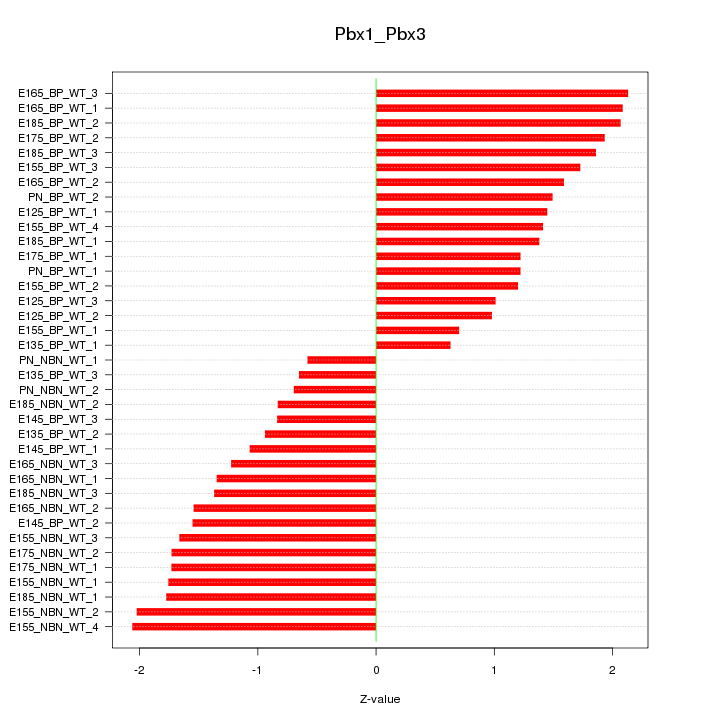
Motif ID: Pbx1_Pbx3
Z-value: 1.465
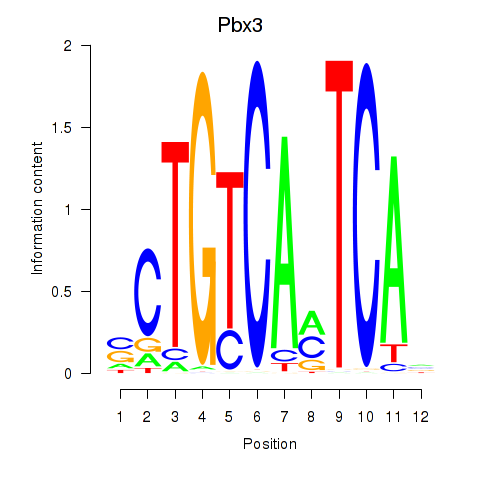
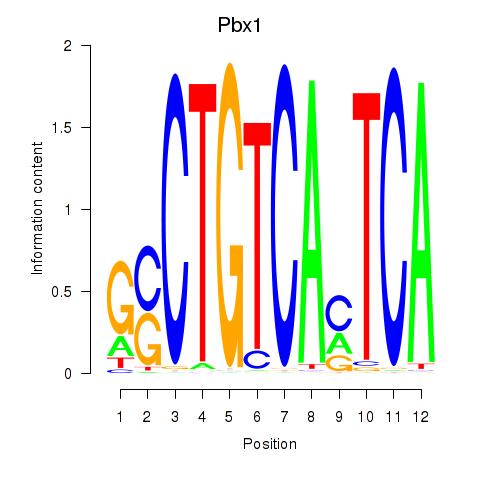
Transcription factors associated with Pbx1_Pbx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pbx1 | ENSMUSG00000052534.9 | Pbx1 |
| Pbx3 | ENSMUSG00000038718.9 | Pbx3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx3 | mm10_v2_chr2_-_34372004_34372044 | 0.74 | 1.3e-07 | Click! |
| Pbx1 | mm10_v2_chr1_-_168432270_168432347 | -0.54 | 6.0e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 2.7 | 8.2 | GO:0060023 | soft palate development(GO:0060023) |
| 2.1 | 16.6 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 1.3 | 7.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.2 | 5.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.2 | 9.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 1.1 | 3.4 | GO:0072554 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.1 | 3.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.0 | 3.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.9 | 2.7 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.8 | 2.5 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.8 | 2.3 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.8 | 2.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.8 | 16.6 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.7 | 11.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 2.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.6 | 1.9 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.6 | 1.9 | GO:0019230 | proprioception(GO:0019230) |
| 0.6 | 21.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.5 | 4.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.5 | 2.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.5 | 6.9 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.5 | 2.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.5 | 3.3 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.5 | 3.8 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.5 | 2.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 1.7 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.4 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.7 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.3 | 0.7 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.3 | 0.3 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.3 | 2.0 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.3 | 1.6 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.3 | 0.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 1.9 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.3 | 1.3 | GO:0001692 | histamine metabolic process(GO:0001692) histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 | 2.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 2.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 3.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 4.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 | 3.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.3 | 1.3 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.3 | 1.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.7 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.2 | 0.9 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 4.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.2 | 0.9 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.2 | 2.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.6 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.2 | 2.7 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 2.8 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 0.4 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.2 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.5 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.2 | 3.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 1.0 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 0.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.9 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.4 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.3 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 2.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 2.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 1.0 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.3 | GO:0070973 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.3 | GO:1904207 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 1.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.2 | GO:0060047 | heart process(GO:0003015) heart contraction(GO:0060047) |
| 0.1 | 0.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 2.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 2.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.6 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.0 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 2.4 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 0.8 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.6 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 3.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:1903756 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 2.4 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 3.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.8 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 5.1 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 2.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 4.4 | GO:0048469 | cell maturation(GO:0048469) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 4.3 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 2.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 2.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 8.6 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 2.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 16.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 1.6 | 4.8 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 2.8 | GO:0008623 | CHRAC(GO:0008623) |
| 0.6 | 7.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 3.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 2.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 3.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 2.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 3.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 8.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 15.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 3.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 6.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0030670 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.3 | 3.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.2 | 17.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.1 | 3.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 6.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 16.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.8 | 3.1 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.6 | 2.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.6 | 4.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 3.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 2.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 2.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 3.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.7 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 2.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 0.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 7.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 2.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 7.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 7.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 2.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 4.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 3.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 13.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 1.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 3.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.4 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.3 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 3.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.4 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 18.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 3.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.8 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 9.7 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 0.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.4 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 11.1 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.2 | 14.9 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.2 | 6.6 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 8.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.5 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.0 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 3.4 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 6.9 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 2.4 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.9 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.8 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 1.5 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 1.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 1.5 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 2.9 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 7.3 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.4 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 3.3 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 4.4 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 2.2 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 10.5 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.1 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.3 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.4 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.9 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.7 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.9 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.2 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.8 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |