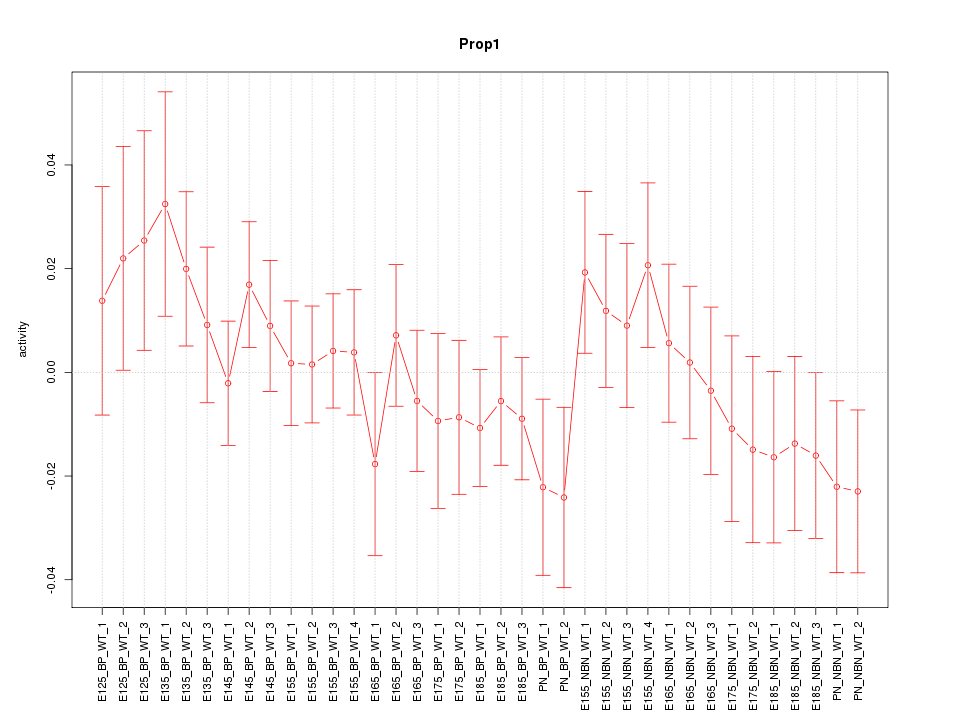
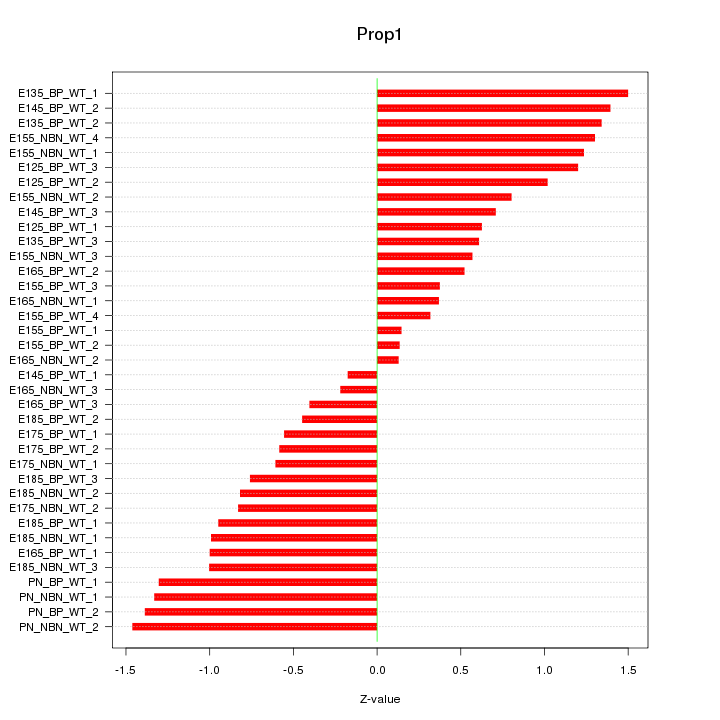
Motif ID: Prop1
Z-value: 0.892
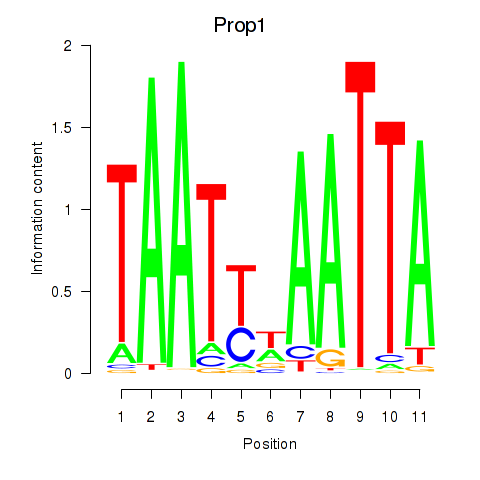
Transcription factors associated with Prop1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Prop1 | ENSMUSG00000044542.3 | Prop1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prop1 | mm10_v2_chr11_-_50953745_50953768 | 0.01 | 9.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.2 | 5.9 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.8 | 4.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.8 | 11.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.7 | 2.9 | GO:1900623 | positive regulation of keratinocyte proliferation(GO:0010838) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.7 | 6.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.7 | 3.3 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.6 | 4.6 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.5 | 2.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.5 | 2.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 1.4 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 | 2.3 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.4 | 1.3 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.4 | 5.5 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.4 | 1.1 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.3 | 1.6 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.3 | 1.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.3 | 1.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 3.7 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.3 | 1.7 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 1.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 2.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.2 | 2.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.1 | GO:1902995 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.6 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 0.6 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 | 1.4 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 1.2 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 1.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 0.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 2.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 7.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.4 | GO:2000318 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 1.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 2.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 1.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 3.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.4 | GO:1903587 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.3 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.4 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.8 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 2.4 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.1 | GO:2000338 | positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial RNA 3'-end processing(GO:0000965) mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 1.1 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.6 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.0 | 3.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 2.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0060161 | histone H4-R3 methylation(GO:0043985) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 3.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 2.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 1.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 1.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 2.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 1.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.3 | 1.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 3.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.9 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.4 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 3.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 11.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 1.2 | 4.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 2.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 8.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.8 | 2.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 2.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.7 | 2.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.5 | 1.6 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.4 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 1.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 0.9 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.3 | 0.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 2.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.5 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 10.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 5.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 11.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 3.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 3.6 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 3.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.5 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 3.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.1 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.9 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 9.4 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 8.2 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.3 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.0 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.6 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 3.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.2 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.7 | 2.1 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 2.5 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 2.9 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 1.6 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 3.5 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 5.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.9 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.2 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.1 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.5 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.4 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.6 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.6 | REACTOME_AQUAPORIN_MEDIATED_TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 2.1 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 2.5 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.0 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |