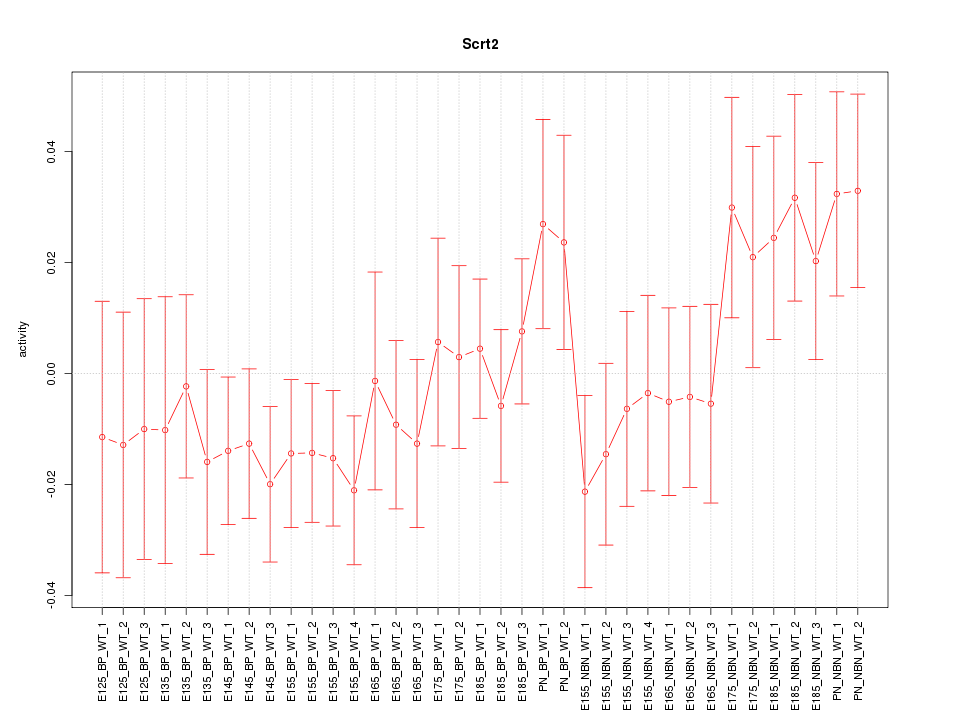
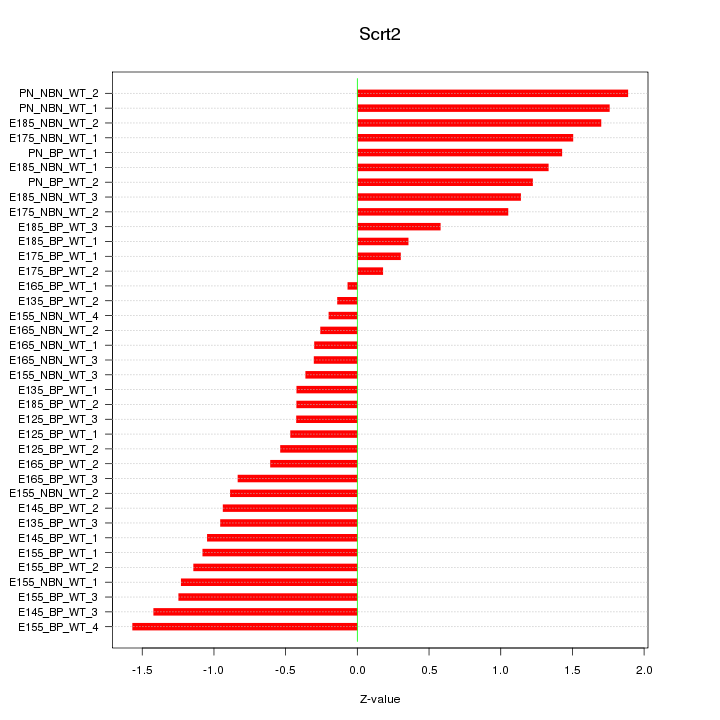
Motif ID: Scrt2
Z-value: 0.992
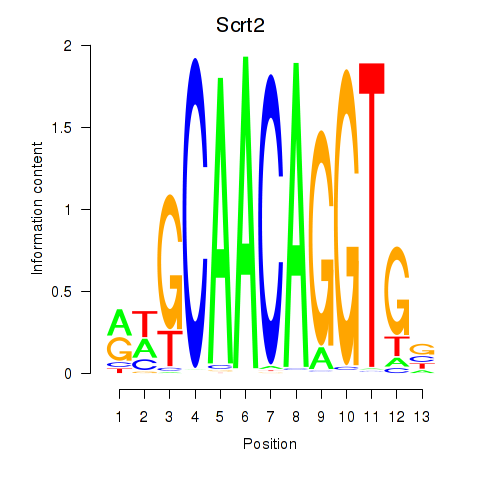
Transcription factors associated with Scrt2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Scrt2 | ENSMUSG00000060257.2 | Scrt2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | mm10_v2_chr2_+_152081529_152081624 | -0.62 | 4.7e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 1.1 | 8.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 1.0 | 3.0 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.9 | 4.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.9 | 3.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 3.4 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.6 | 1.9 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.5 | 2.3 | GO:0015822 | ornithine transport(GO:0015822) |
| 0.4 | 1.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 0.9 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.3 | 1.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 1.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 3.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 1.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 1.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.6 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 0.2 | 2.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 1.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 1.2 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.1 | 1.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 1.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 4.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 3.1 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 3.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 4.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 2.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.3 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.3 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.4 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 3.1 | GO:0031102 | neuron projection regeneration(GO:0031102) response to axon injury(GO:0048678) |
| 0.1 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 1.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 1.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 1.9 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.3 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 4.6 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) response to cAMP(GO:0051591) |
| 0.0 | 0.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 1.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 3.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 7.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 7.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 7.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 3.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.8 | 3.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 1.9 | GO:0019002 | GMP binding(GO:0019002) |
| 0.6 | 2.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.3 | 3.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 3.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 0.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 6.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 1.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 4.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 5.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 3.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 4.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 3.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 4.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 4.6 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 11.9 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.9 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.6 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 7.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 1.7 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.5 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.2 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.9 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.3 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.3 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.4 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.9 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.4 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.2 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.8 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |