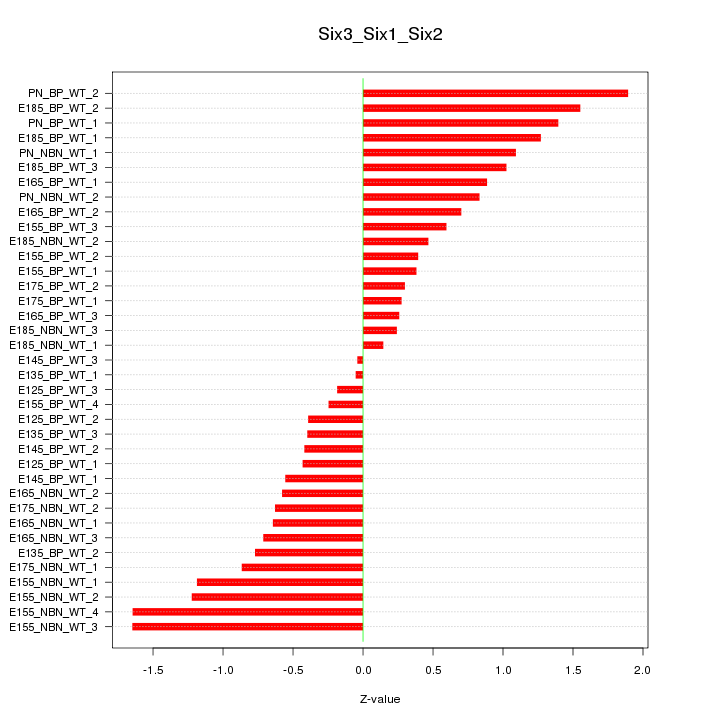
Motif ID: Six3_Six1_Six2
Z-value: 0.860
Transcription factors associated with Six3_Six1_Six2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Six1 | ENSMUSG00000051367.8 | Six1 |
| Six2 | ENSMUSG00000024134.10 | Six2 |
| Six3 | ENSMUSG00000038805.9 | Six3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six1 | mm10_v2_chr12_-_73047179_73047179 | 0.46 | 4.0e-03 | Click! |
| Six2 | mm10_v2_chr17_-_85688252_85688275 | 0.32 | 5.4e-02 | Click! |
| Six3 | mm10_v2_chr17_+_85621017_85621031 | 0.11 | 5.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:1904706 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.8 | 4.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.6 | 6.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.6 | 1.7 | GO:0006097 | glyoxylate cycle(GO:0006097) negative regulation of glial cell migration(GO:1903976) |
| 0.3 | 2.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 2.6 | GO:0036159 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.2 | 0.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 2.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.5 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.0 | 0.1 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 1.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 1.2 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 3.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 1.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 0.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 5.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.4 | GO:0000776 | kinetochore(GO:0000776) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.6 | 3.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.6 | 1.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 0.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 2.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 2.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 4.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.5 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |