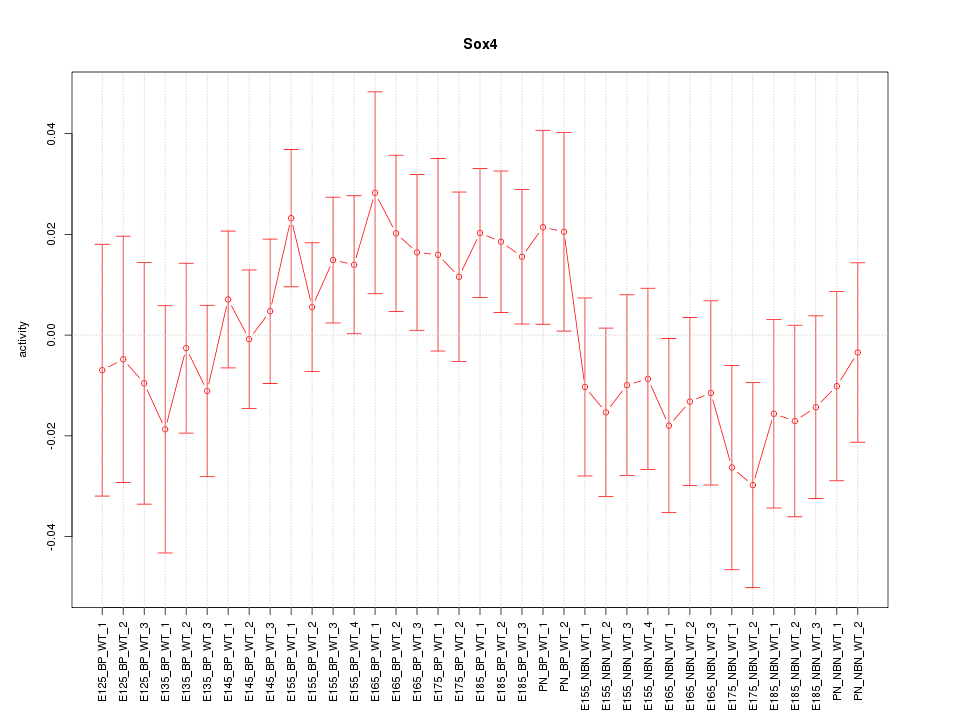
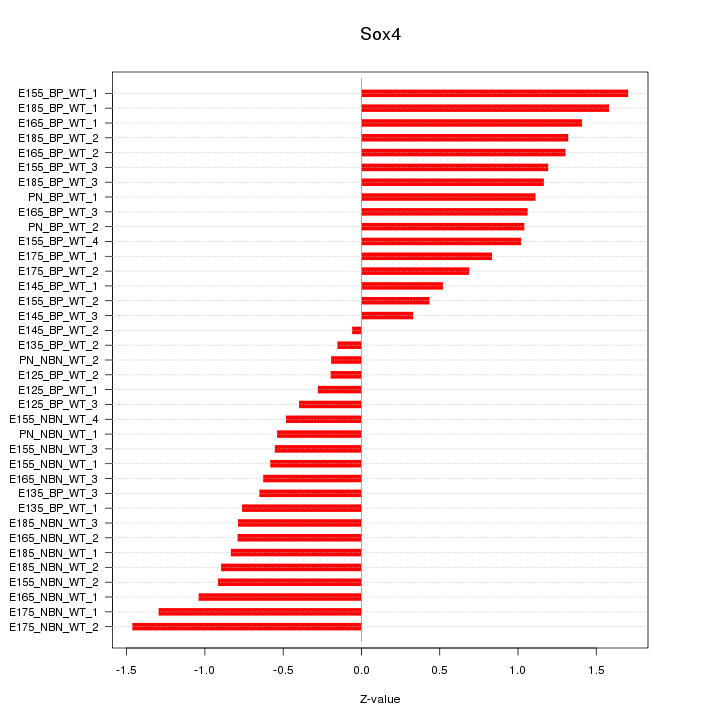
Motif ID: Sox4
Z-value: 0.919

Transcription factors associated with Sox4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox4 | ENSMUSG00000076431.4 | Sox4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox4 | mm10_v2_chr13_-_28953690_28953713 | 0.38 | 2.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.8 | 2.4 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.7 | 2.9 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.6 | 2.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.6 | 7.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.6 | 1.8 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.5 | 1.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 | 3.0 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.3 | 0.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 1.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 3.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 5.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 0.7 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 5.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 3.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 5.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 2.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.9 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 1.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.9 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 1.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 2.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 2.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 3.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.8 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 1.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 2.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.8 | 5.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.4 | 5.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 3.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 1.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 0.9 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 2.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 1.0 | GO:0031090 | organelle membrane(GO:0031090) |
| 0.1 | 1.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 2.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 6.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0071004 | U2 snRNP(GO:0005686) U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.8 | 7.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.8 | 3.1 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.6 | 2.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 2.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 5.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 1.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 3.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 2.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 3.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 5.6 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.0 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 7.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.0 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.5 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID_ATR_PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.6 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 1.6 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 5.3 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.9 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.9 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.0 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.2 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |