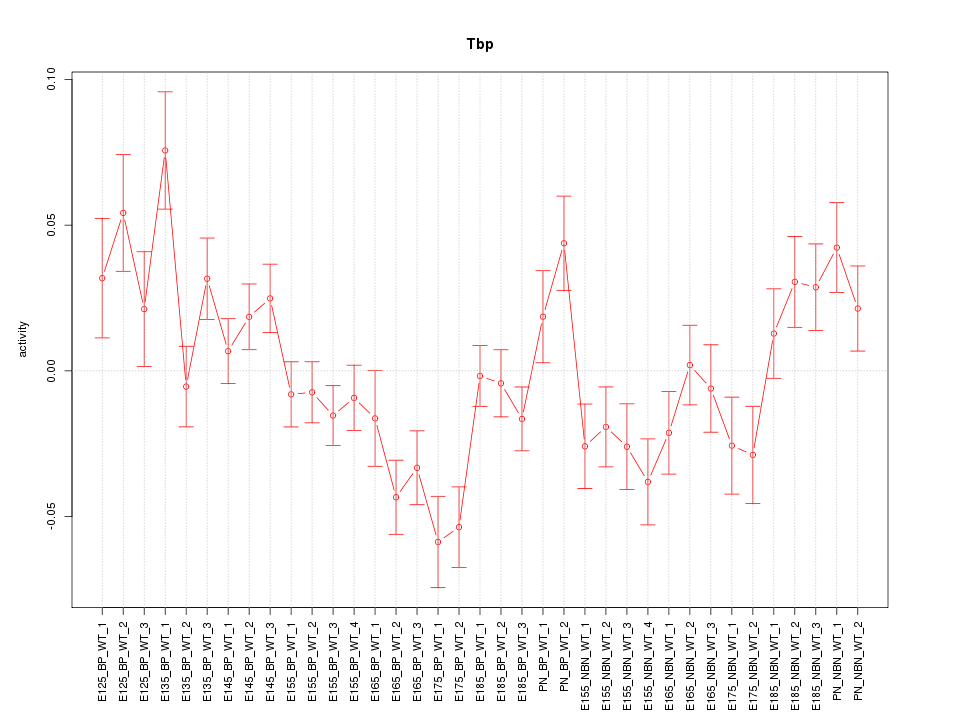
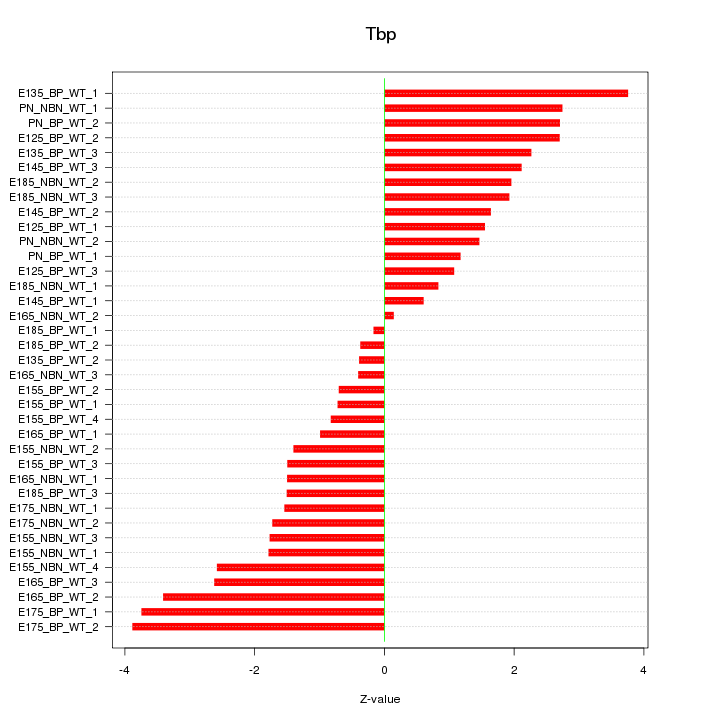
Motif ID: Tbp
Z-value: 1.961
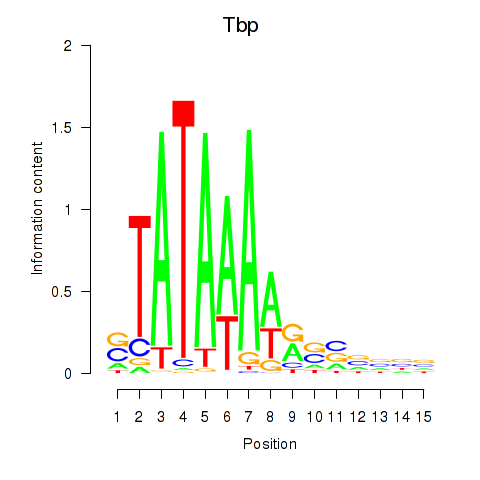
Transcription factors associated with Tbp:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbp | ENSMUSG00000014767.10 | Tbp |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | mm10_v2_chr17_+_15499888_15499960 | -0.44 | 6.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.9 | 5.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.6 | 4.8 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.5 | 7.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 1.3 | 5.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.3 | 5.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.1 | 4.4 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 1.1 | 14.0 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 1.1 | 3.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.0 | 30.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.9 | 2.7 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.9 | 4.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.9 | 2.6 | GO:1990523 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) bone regeneration(GO:1990523) |
| 0.8 | 5.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.8 | 5.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.8 | 2.5 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.8 | 2.3 | GO:1902022 | ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of activation of Janus kinase activity(GO:0010533) L-lysine transport(GO:1902022) |
| 0.7 | 2.2 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.7 | 2.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.7 | 3.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.6 | 2.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.6 | 3.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.6 | 7.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.6 | 4.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.6 | 1.7 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.6 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.5 | 3.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.5 | 1.6 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.5 | 1.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.5 | 2.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.5 | 9.0 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.5 | 2.8 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.4 | 3.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 1.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.4 | 1.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.4 | 2.9 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.4 | 4.1 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.4 | 2.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.4 | 1.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.4 | 1.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.4 | 1.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 6.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 2.6 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.4 | 1.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 1.4 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.4 | 0.7 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.3 | 2.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 1.9 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 2.6 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 | 1.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 1.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 0.9 | GO:0043031 | regulation of phosphatidylcholine catabolic process(GO:0010899) positive regulation of lipoprotein particle clearance(GO:0010986) negative regulation of macrophage activation(GO:0043031) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) regulation of microglial cell activation(GO:1903978) |
| 0.3 | 4.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.3 | 1.1 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 0.8 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.3 | 0.8 | GO:0042713 | sperm ejaculation(GO:0042713) negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 2.1 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 0.7 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.9 | GO:1904996 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.2 | 16.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 1.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 0.8 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 4.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.9 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.2 | 1.2 | GO:0021886 | female meiosis I(GO:0007144) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 0.5 | GO:0097026 | myeloid dendritic cell chemotaxis(GO:0002408) dendritic cell dendrite assembly(GO:0097026) mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 1.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.4 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 1.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 2.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.3 | GO:1903225 | regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 1.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 2.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 1.4 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 3.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.9 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.1 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 | 0.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.5 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.6 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.1 | 1.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 3.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 1.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 3.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 1.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.1 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.8 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 1.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 1.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.5 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.8 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 1.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 5.2 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 2.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.6 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 2.5 | GO:0007283 | spermatogenesis(GO:0007283) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.7 | 5.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.1 | 35.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.0 | 8.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.9 | 2.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.8 | 3.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.8 | 5.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 3.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 16.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 10.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.4 | 1.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 2.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 1.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 2.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 3.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.1 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.2 | 5.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 4.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 0.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 0.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 7.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 2.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 6.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 10.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 2.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 2.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.5 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 7.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.5 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 2.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 14.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.1 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 3.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.3 | 14.0 | GO:0019841 | retinol binding(GO:0019841) |
| 1.1 | 4.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.1 | 9.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.0 | 5.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.9 | 2.7 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.8 | 2.5 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.8 | 8.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.8 | 3.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 2.0 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.6 | 3.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.6 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 25.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.5 | 2.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 1.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.4 | 1.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 2.9 | GO:0032564 | dATP binding(GO:0032564) |
| 0.3 | 1.9 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 1.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 1.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 2.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 7.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 0.8 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.3 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 7.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 6.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 1.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 5.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0097493 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 4.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 2.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 4.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 6.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 14.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 7.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 2.2 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 31.4 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.2 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.2 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 11.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.2 | 8.0 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 1.1 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 2.2 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 4.9 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 5.0 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.5 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.9 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.4 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 8.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 4.3 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID_LYMPH_ANGIOGENESIS_PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.7 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.3 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.4 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.9 | 46.2 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 2.5 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 1.4 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.5 | 5.2 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 5.1 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 2.4 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 6.6 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.2 | 6.6 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 1.7 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 1.2 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 6.0 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.1 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.1 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 7.3 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.1 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.1 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.0 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 6.6 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.1 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.3 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.9 | REACTOME_LIPOPROTEIN_METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 1.0 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.8 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 8.6 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.3 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.1 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.9 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.3 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.0 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.8 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.4 | REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.8 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.7 | REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 3.2 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.1 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.0 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |