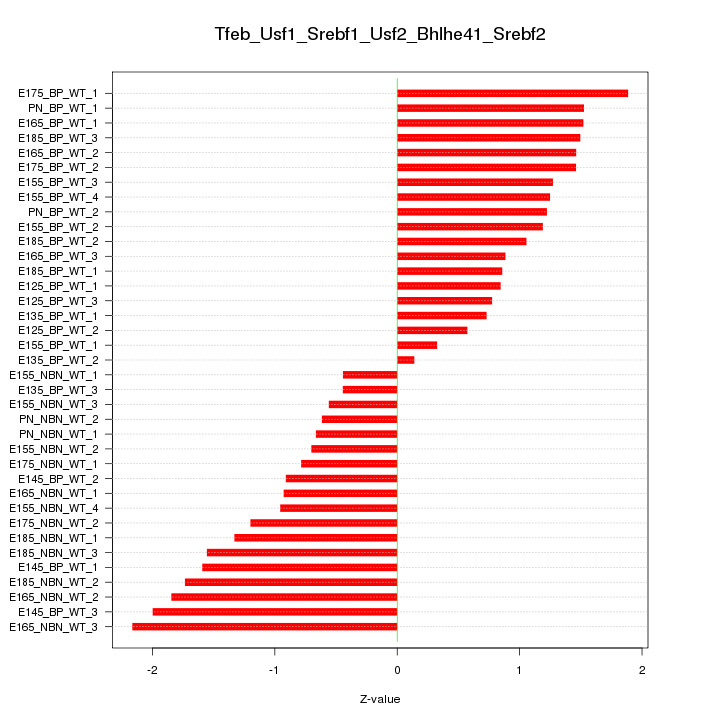
Motif ID: Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Z-value: 1.211
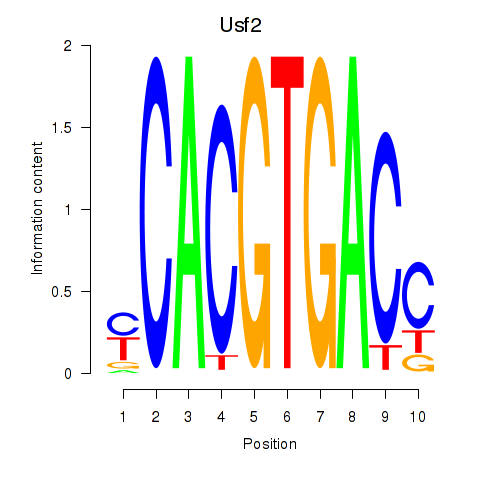
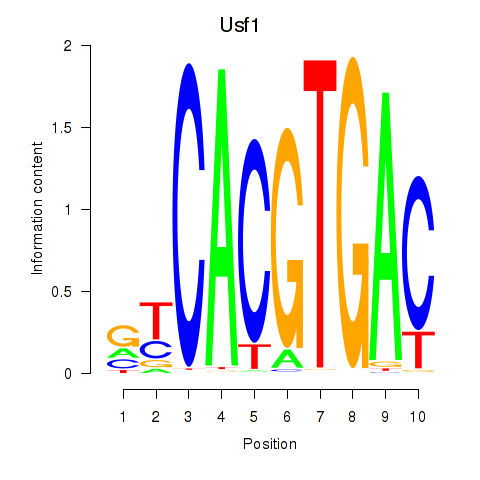
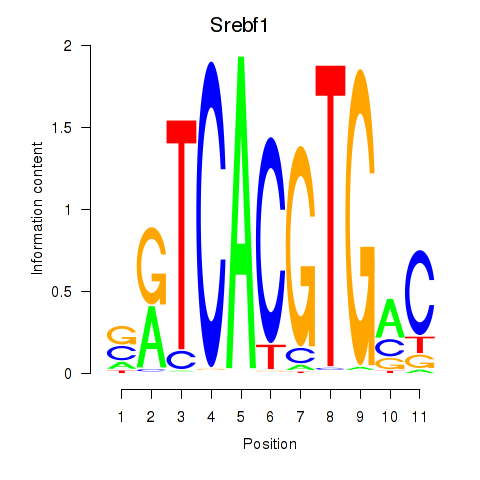
Transcription factors associated with Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bhlhe41 | ENSMUSG00000030256.5 | Bhlhe41 |
| Srebf1 | ENSMUSG00000020538.9 | Srebf1 |
| Srebf2 | ENSMUSG00000022463.7 | Srebf2 |
| Tfeb | ENSMUSG00000023990.12 | Tfeb |
| Usf1 | ENSMUSG00000026641.7 | Usf1 |
| Usf2 | ENSMUSG00000058239.7 | Usf2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srebf1 | mm10_v2_chr11_-_60220550_60220632 | 0.69 | 2.6e-06 | Click! |
| Tfeb | mm10_v2_chr17_+_47785720_47785739 | 0.52 | 1.1e-03 | Click! |
| Srebf2 | mm10_v2_chr15_+_82147238_82147275 | -0.35 | 3.5e-02 | Click! |
| Usf1 | mm10_v2_chr1_+_171411343_171411384 | 0.24 | 1.6e-01 | Click! |
| Usf2 | mm10_v2_chr7_-_30956742_30956803 | -0.21 | 2.1e-01 | Click! |
First level regulatory network of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
View svg image
View png image
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 21.0 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 4.4 | 17.7 | GO:0010288 | response to lead ion(GO:0010288) |
| 3.3 | 9.8 | GO:0030421 | defecation(GO:0030421) |
| 2.9 | 8.7 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 2.8 | 8.3 | GO:1904959 | elastin biosynthetic process(GO:0051542) copper ion export(GO:0060003) regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.7 | 13.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 2.3 | 6.9 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 2.3 | 9.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 2.2 | 8.9 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 2.2 | 8.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.7 | 5.0 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 1.5 | 7.6 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.4 | 5.4 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 1.3 | 3.9 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 1.3 | 3.8 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.3 | 5.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.2 | 3.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.2 | 7.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.1 | 5.4 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 1.1 | 10.6 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 1.0 | 3.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 1.0 | 3.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.0 | 3.0 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.0 | 1.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.0 | 11.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.9 | 3.8 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.9 | 5.6 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.9 | 4.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.9 | 5.4 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.9 | 2.7 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.9 | 2.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.8 | 4.9 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.8 | 1.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.8 | 3.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.8 | 5.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.8 | 3.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 0.7 | 4.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.7 | 3.5 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.7 | 1.3 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.6 | 3.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.6 | 5.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.6 | 1.9 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.6 | 6.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.6 | 1.7 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.6 | 1.7 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 | 2.8 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.5 | 2.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.5 | 2.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.5 | 3.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.5 | 3.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 1.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.5 | 1.9 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.5 | 2.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.5 | 1.4 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.5 | 5.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.5 | 3.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 3.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 1.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 2.6 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.4 | 2.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.4 | 1.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 1.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.2 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.4 | 2.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.4 | 3.4 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.4 | 2.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.4 | 1.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.4 | 1.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.4 | 3.6 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.4 | 3.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.4 | 2.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 1.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.3 | 1.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 4.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.3 | 2.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 2.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 18.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 1.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 0.9 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.3 | 0.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 4.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 | 2.5 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.3 | 0.9 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.3 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 3.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 1.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 1.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.3 | 6.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.3 | 0.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 0.9 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 | 1.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 6.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.3 | 2.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.3 | 1.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 1.3 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.3 | 2.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 0.8 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 15.2 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.3 | 3.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 0.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.3 | 1.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 3.4 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 2.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 1.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.1 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.2 | 2.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.7 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.2 | 0.7 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.2 | 2.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 3.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.9 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 2.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 2.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 2.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 1.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.2 | 0.8 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 0.8 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 0.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 0.4 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 2.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 3.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 3.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 1.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 1.7 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 1.7 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 1.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 0.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 10.2 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.2 | 0.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 0.5 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.2 | 0.8 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.2 | 2.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 0.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 3.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.5 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.2 | 3.6 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.2 | 3.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.5 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.2 | 1.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.5 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.2 | 2.3 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 | 2.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 4.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 5.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 1.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 3.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 1.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 1.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 2.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 10.0 | GO:0043473 | pigmentation(GO:0043473) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.4 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 1.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 2.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.3 | GO:0021972 | corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 3.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.8 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 1.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 1.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 6.2 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 1.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.7 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 1.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.9 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 6.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.6 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 1.1 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 2.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 8.6 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 4.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 1.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 1.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.4 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.1 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.1 | GO:0035826 | rubidium ion transport(GO:0035826) |
| 0.1 | 0.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 3.0 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.4 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 0.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 0.4 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.4 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 1.1 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.2 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 0.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.2 | GO:0009946 | optic vesicle morphogenesis(GO:0003404) proximal/distal axis specification(GO:0009946) neuroblast migration(GO:0097402) |
| 0.1 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.1 | 0.8 | GO:0043487 | regulation of RNA stability(GO:0043487) |
| 0.1 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.2 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 1.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.4 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.4 | GO:0019359 | NAD biosynthetic process(GO:0009435) nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.1 | 1.2 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.1 | 0.4 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.8 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.1 | 0.8 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.8 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.1 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 1.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 2.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.3 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.1 | 0.2 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.5 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.2 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 1.2 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.6 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 0.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.2 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.6 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 2.2 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.3 | GO:0035561 | regulation of chromatin binding(GO:0035561) |
| 0.0 | 1.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.4 | GO:0010664 | negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.6 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.9 | GO:0001889 | liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 1.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.6 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0045911 | positive regulation of DNA recombination(GO:0045911) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) dibasic protein processing(GO:0090472) negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:1903364 | positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.0 | 0.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.0 | 0.0 | GO:0071865 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) apoptotic process in bone marrow(GO:0071839) regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.0 | 0.1 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.0 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0061180 | mammary gland epithelium development(GO:0061180) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.5 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.4 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 1.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 3.0 | 11.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 2.5 | 7.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.7 | 8.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.7 | 6.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.2 | 4.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 5.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.0 | 6.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.8 | 1.7 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.8 | 2.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.7 | 9.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.7 | 3.0 | GO:1902737 | dendritic filopodium(GO:1902737) growth cone filopodium(GO:1990812) |
| 0.7 | 2.9 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.7 | 5.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 5.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 9.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 2.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.6 | 1.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.6 | 1.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.5 | 2.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 3.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 3.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 2.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 1.3 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.4 | 6.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.4 | 1.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 3.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 3.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 3.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 6.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.4 | 1.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 3.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 1.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 1.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 1.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 1.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 6.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 2.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 3.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 5.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 2.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 8.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 1.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 3.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 2.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.3 | 13.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 3.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 1.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 1.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 5.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 1.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 0.9 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 6.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 3.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 1.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 4.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 3.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 3.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 3.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.7 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 2.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 10.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.1 | 4.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 2.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.2 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 2.6 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 4.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 3.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 4.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 12.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.0 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 2.8 | 8.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.6 | 4.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.6 | 10.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.3 | 7.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.2 | 5.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.1 | 3.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 1.0 | 4.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.0 | 3.0 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.9 | 3.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.9 | 2.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.8 | 3.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.8 | 7.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.7 | 5.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.7 | 2.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.7 | 2.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 4.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 4.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.6 | 2.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.6 | 1.8 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.6 | 3.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.5 | 3.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.5 | 5.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.5 | 3.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 3.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 2.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.5 | 1.5 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.5 | 6.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 3.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.5 | 3.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 3.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 0.5 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 1.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.4 | 3.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 2.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 19.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 1.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 0.4 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 2.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.4 | 14.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 1.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.7 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.3 | 2.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 0.9 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 3.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.3 | 1.2 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.3 | 2.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 0.9 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 1.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 1.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 1.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.0 | GO:0004844 | mismatch base pair DNA N-glycosylase activity(GO:0000700) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 2.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 1.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 3.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 1.7 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.2 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 4.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.6 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.2 | 1.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 2.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 1.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 0.8 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 4.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.2 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 9.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 1.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 9.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 1.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.2 | 3.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.5 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 7.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 0.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 1.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 3.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 3.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.5 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.4 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 37.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 5.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.8 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 7.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 38.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 1.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 3.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 2.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 3.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 3.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.2 | GO:0015116 | sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 5.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 2.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 9.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 2.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 3.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 0.5 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 3.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.8 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 3.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 1.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 4.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 4.1 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 3.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.3 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 3.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 4.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.3 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.0 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0016830 | carbon-carbon lyase activity(GO:0016830) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 18.9 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 4.8 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 14.1 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.3 | 12.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.3 | 2.2 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 7.0 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.3 | 7.8 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 1.4 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.5 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 6.5 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 5.0 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.8 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.6 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.0 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.5 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 2.7 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.3 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.7 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.2 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.8 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.0 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.7 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.6 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.0 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.4 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 0.2 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 0.3 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.0 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.1 | 1.6 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 0.4 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.9 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.4 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.1 | 4.3 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.4 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 3.6 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.2 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 4.8 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.2 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.0 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 15.4 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 4.9 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 3.6 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 19.1 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.4 | 6.7 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.4 | 5.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.3 | 8.9 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 18.2 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 4.7 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.3 | 1.8 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 2.8 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 3.0 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 0.9 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 7.8 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 3.2 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 3.4 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 4.3 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 4.7 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 0.5 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 3.6 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 5.3 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.6 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_MULTIPLE_NUCLEOTIDE_PATCH_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.1 | 1.1 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.2 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 9.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.9 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 2.8 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 11.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.3 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 5.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 4.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.7 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.5 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.8 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 11.5 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 14.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.5 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.6 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.6 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.2 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.4 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.4 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.0 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.6 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 0.3 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.8 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.4 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.3 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.0 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.1 | REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 7.3 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME_ION_CHANNEL_TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 2.4 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.7 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.9 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME_DNA_REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.1 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |