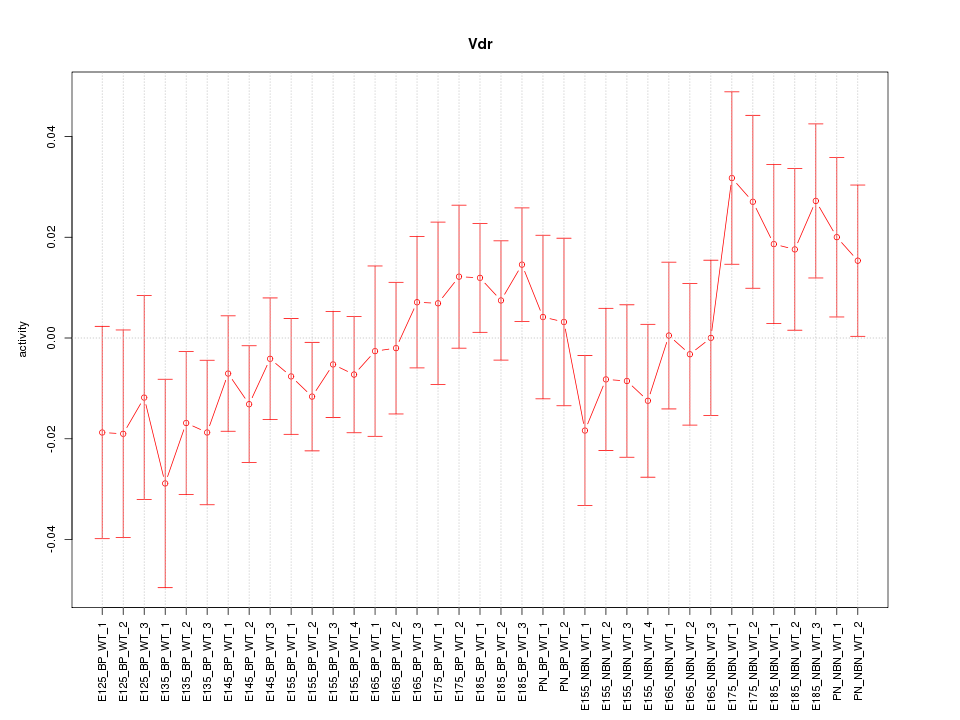
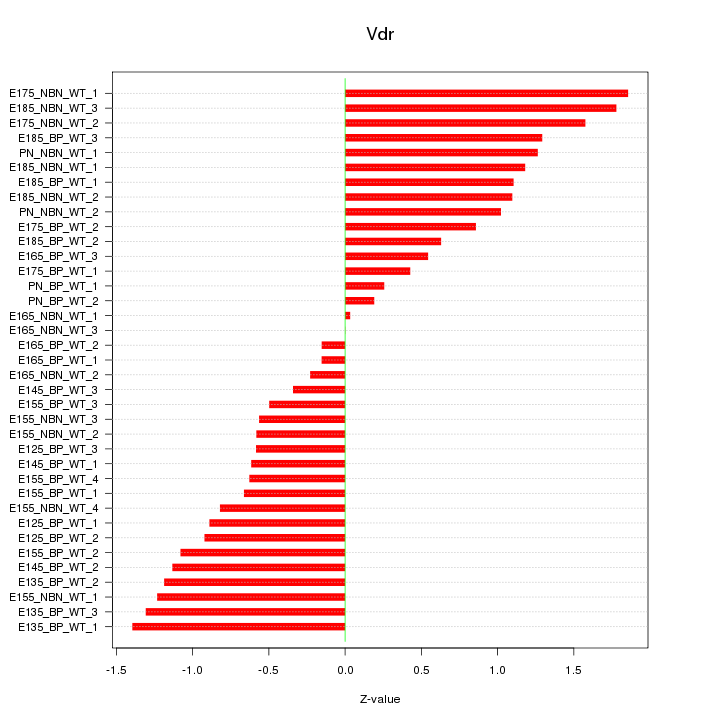
Motif ID: Vdr
Z-value: 0.944
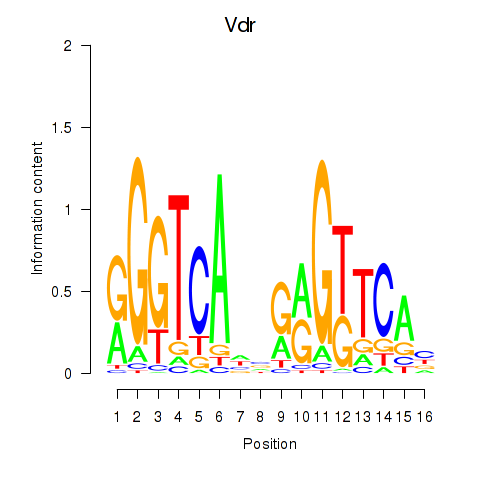
Transcription factors associated with Vdr:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Vdr | ENSMUSG00000022479.9 | Vdr |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 1.2 | 3.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.1 | 3.4 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.9 | 2.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.8 | 3.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.8 | 4.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.8 | 8.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.8 | 4.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.8 | 3.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 1.4 | GO:1902022 | L-lysine transport(GO:1902022) |
| 0.6 | 0.6 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.5 | 2.4 | GO:0015817 | glutamine transport(GO:0006868) histidine transport(GO:0015817) cellular response to potassium ion starvation(GO:0051365) |
| 0.5 | 1.4 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.4 | 1.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.3 | 1.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 0.8 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 0.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 2.8 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.2 | 0.6 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.6 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 | 1.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.6 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 1.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 2.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 1.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 2.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.3 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.1 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 3.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 1.3 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 1.0 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.0 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.5 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 2.6 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.8 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.6 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.0 | 1.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.7 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.0 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.0 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.7 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.0 | 4.0 | GO:0097487 | vesicle lumen(GO:0031983) multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 2.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.4 | 2.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 1.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 1.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 3.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 0.9 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.2 | 0.8 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.8 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.2 | 1.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.8 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 8.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 3.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.3 | 4.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.2 | 3.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 1.1 | 3.4 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.8 | 2.4 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.7 | 8.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.5 | 4.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 1.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.4 | 1.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.4 | 4.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 1.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 3.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 2.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 0.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.6 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.2 | 1.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.5 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 2.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 2.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.7 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 2.9 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.4 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 5.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.5 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.3 | NABA_CORE_MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.6 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 1.1 | 8.0 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.9 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 6.6 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 4.0 | REACTOME_SHC1_EVENTS_IN_EGFR_SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 2.2 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.7 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.8 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.9 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.7 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.2 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.5 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.8 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.3 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.0 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.3 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.8 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |