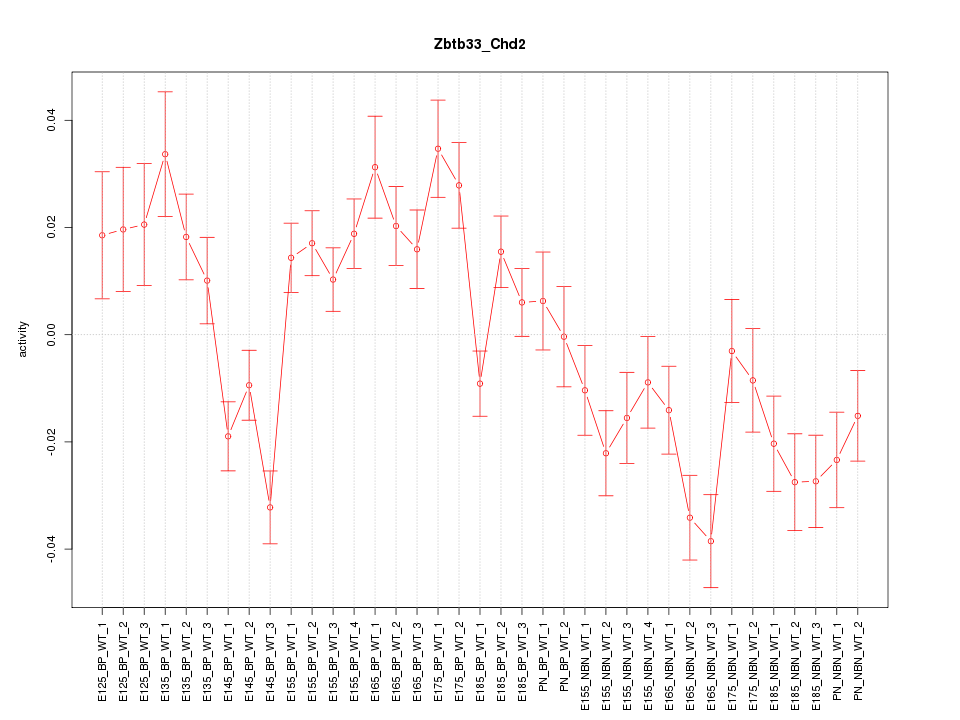
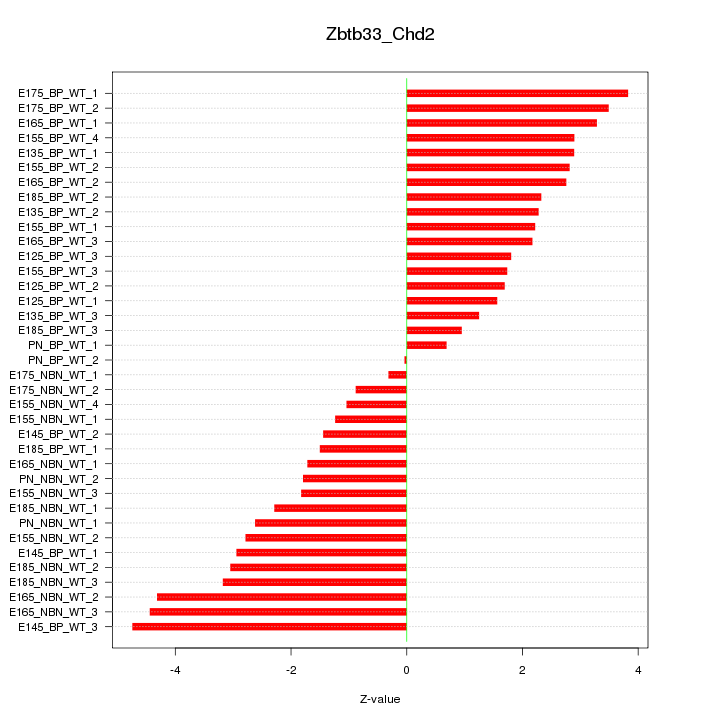
Motif ID: Zbtb33_Chd2
Z-value: 2.498
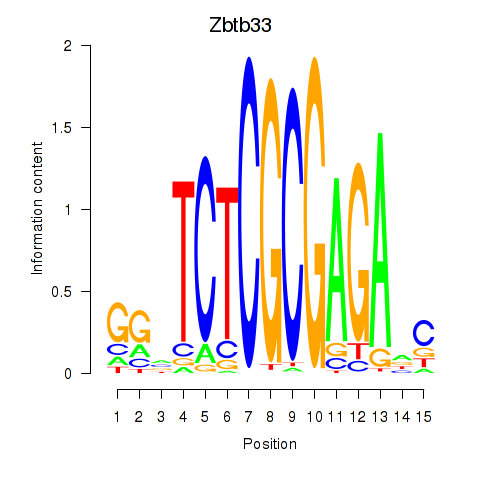
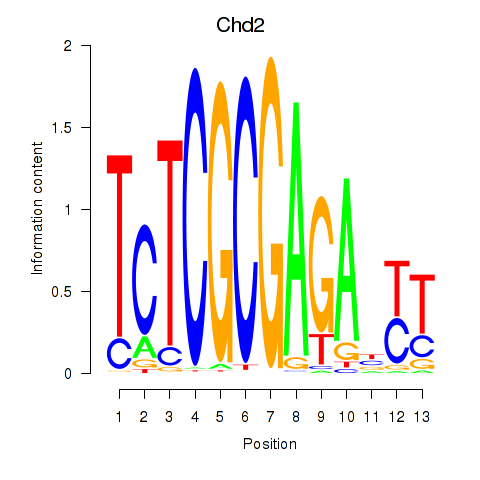
Transcription factors associated with Zbtb33_Chd2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Chd2 | ENSMUSG00000078671.4 | Chd2 |
| Zbtb33 | ENSMUSG00000048047.3 | Zbtb33 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb33 | mm10_v2_chrX_+_38189780_38189826 | 0.84 | 1.1e-10 | Click! |
| Chd2 | mm10_v2_chr7_-_73541738_73541758 | 0.79 | 4.9e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.5 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 2.9 | 8.7 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 2.7 | 8.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 2.5 | 17.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 2.4 | 19.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 2.2 | 6.5 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 1.5 | 4.5 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.5 | 16.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 1.4 | 4.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.3 | 1.3 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 1.2 | 20.9 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 1.2 | 10.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 1.2 | 12.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.2 | 6.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.1 | 3.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 1.1 | 3.4 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 1.1 | 3.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 1.1 | 5.6 | GO:0019230 | proprioception(GO:0019230) |
| 1.0 | 6.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.0 | 4.8 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.8 | 4.2 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.8 | 3.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.8 | 2.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.7 | 2.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.7 | 2.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 2.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.7 | 2.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.6 | 1.9 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.6 | 10.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.6 | 16.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.6 | 1.7 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.6 | 1.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.5 | 1.6 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.5 | 6.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.5 | 9.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.5 | 4.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.5 | 1.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.5 | 2.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.4 | 3.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.4 | 3.0 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 2.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.2 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.4 | 1.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.4 | 3.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.4 | 1.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 | 2.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.4 | 1.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 1.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 1.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 2.4 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.3 | 10.1 | GO:0045005 | DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.3 | 1.7 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.3 | 1.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 1.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.3 | 4.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 0.9 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.3 | 0.9 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.3 | 2.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 1.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.3 | 1.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 0.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 3.0 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.3 | 1.0 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 1.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 1.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 2.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.9 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 3.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 1.3 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 2.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 0.8 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 15.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 8.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 1.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 0.8 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 1.3 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.2 | 1.5 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.2 | 0.7 | GO:0060744 | thelarche(GO:0042695) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) mammary gland branching involved in thelarche(GO:0060744) mammary gland branching involved in pregnancy(GO:0060745) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 1.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 4.0 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 1.7 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.2 | 1.5 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.2 | 2.5 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 0.5 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 2.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 1.0 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 0.5 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.2 | 0.8 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 0.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 3.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.7 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 1.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 1.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 2.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 4.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.9 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 1.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 4.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 9.1 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 | 4.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.3 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 2.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 2.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.4 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 1.1 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.8 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 7.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 2.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.7 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 0.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.4 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 2.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 6.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 0.3 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 2.3 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 0.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.4 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.5 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.8 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 | 0.1 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 1.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.7 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 2.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 5.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 7.1 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 3.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 1.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 1.2 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.0 | 0.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.0 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 0.7 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 2.6 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.5 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.7 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 39.4 | GO:0000796 | condensin complex(GO:0000796) |
| 2.7 | 8.0 | GO:0035101 | FACT complex(GO:0035101) |
| 2.6 | 7.9 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 1.8 | 23.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.7 | 8.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.1 | 3.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.1 | 4.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 1.0 | 4.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.9 | 3.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.9 | 2.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.8 | 2.5 | GO:0005940 | septin ring(GO:0005940) |
| 0.7 | 4.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.7 | 19.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.6 | 6.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 1.8 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.5 | 1.6 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.5 | 2.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 1.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.5 | 2.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.5 | 1.9 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.5 | 11.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.5 | 2.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 12.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.4 | 2.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.4 | 3.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 6.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.4 | 2.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 3.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 2.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 1.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 1.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 1.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 0.9 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 5.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 1.7 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.2 | 0.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 5.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 0.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 1.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 2.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 6.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 4.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 5.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 2.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 6.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.4 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 13.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 6.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 3.6 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 16.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.7 | 8.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.7 | 9.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.6 | 4.9 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.3 | 9.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.8 | 6.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.7 | 2.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.7 | 2.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.6 | 1.8 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.5 | 1.6 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.5 | 2.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 0.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.4 | 2.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 2.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 3.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.4 | 2.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 16.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.4 | 1.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 3.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 7.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 1.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 1.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.3 | 5.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 1.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.3 | 3.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 4.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 2.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 4.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 1.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 3.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 4.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 2.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.3 | 0.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 1.0 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 7.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 0.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 1.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 2.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 8.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 0.8 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 15.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 0.8 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.2 | 4.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.0 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 1.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 4.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 21.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 23.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 6.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 16.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 2.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.5 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 3.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 1.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.4 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 3.4 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 2.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 6.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 13.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 13.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 4.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 3.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 2.9 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 0.0 | 0.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 7.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 5.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 3.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 3.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 40.2 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.7 | 34.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 9.9 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.3 | 8.8 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 12.8 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.2 | 10.3 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 3.6 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.4 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.1 | 11.9 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 8.2 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.0 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.9 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 1.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.7 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.2 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 3.0 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.6 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.7 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 2.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID_LKB1_PATHWAY | LKB1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.4 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.5 | 8.7 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 3.0 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 2.3 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 8.7 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.9 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 18.5 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 5.0 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.2 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 5.0 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.5 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.5 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.4 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 2.7 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.5 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.7 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 1.3 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.2 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.4 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.8 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.2 | REACTOME_RORA_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 1.4 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.6 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 0.8 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 3.1 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.7 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 3.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.3 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.4 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 10.6 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME_GLOBAL_GENOMIC_NER_GG_NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 2.7 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 3.2 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.9 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |