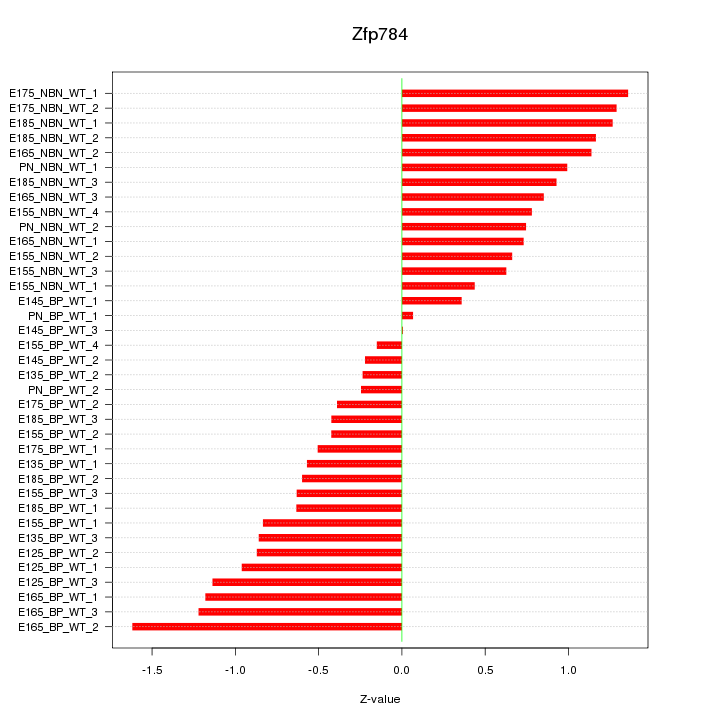
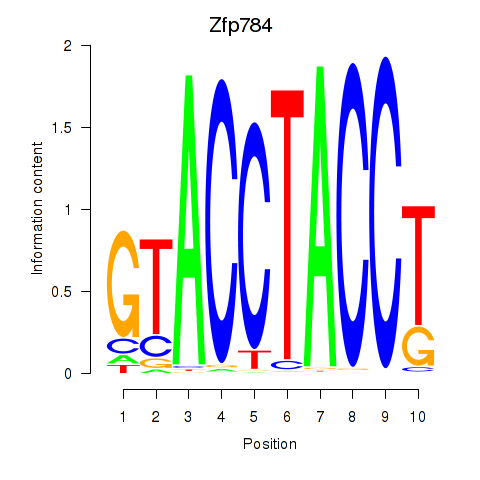
Motif ID: Zfp784
Z-value: 0.831
Transcription factors associated with Zfp784:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp784 | ENSMUSG00000043290.6 | Zfp784 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp784 | mm10_v2_chr7_-_5038427_5038450 | -0.01 | 9.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.5 | 4.6 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) negative regulation of lymphocyte migration(GO:2000402) |
| 1.1 | 4.2 | GO:0035793 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 1.0 | 3.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.9 | 3.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.9 | 4.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 3.5 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.8 | 4.6 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.7 | 7.3 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.5 | 2.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 1.6 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.5 | 2.7 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.5 | 2.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 4.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 4.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.4 | 1.2 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.4 | 1.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.4 | 3.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 0.7 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.3 | 1.3 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.3 | 1.0 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.3 | 1.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 1.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 1.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 1.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 0.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 | 0.9 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 2.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 0.8 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.3 | 1.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 3.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 0.5 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 | 2.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 2.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 3.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 3.0 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 1.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 1.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 1.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 14.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 0.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 1.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.2 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 | 0.4 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 1.1 | GO:0014013 | regulation of gliogenesis(GO:0014013) |
| 0.1 | 2.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.0 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.1 | 0.4 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 0.9 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.8 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.8 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.5 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 5.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.4 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 2.7 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.2 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 6.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 3.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 5.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.0 | 0.6 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.4 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 1.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0098903 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.0 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 3.0 | GO:0006869 | lipid transport(GO:0006869) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.9 | 2.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.7 | 3.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.5 | 7.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 1.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.4 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 1.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.4 | 1.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 3.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 5.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.5 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 2.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 16.5 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 2.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 4.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 6.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 4.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.3 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 2.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 1.0 | 3.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.9 | 3.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.6 | 1.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.5 | 2.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 3.2 | GO:1904315 | GABA-gated chloride ion channel activity(GO:0022851) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 2.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.5 | 9.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 3.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 4.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 2.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 1.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 1.3 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.3 | 1.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 1.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 1.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 1.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 3.4 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 10.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 0.8 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 3.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.9 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 2.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 4.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 4.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.2 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 3.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 5.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 3.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 2.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 3.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 3.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 8.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.9 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.2 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 1.5 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.4 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.7 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.5 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 1.5 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 3.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.6 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 3.6 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.3 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 4.4 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 2.4 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 1.3 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.7 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.0 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 3.2 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.0 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.9 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 3.6 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.7 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.9 | REACTOME_PD1_SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.8 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.9 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.1 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.9 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.5 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.1 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.1 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 3.2 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.1 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.9 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.5 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.8 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.3 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.2 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 1.2 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |