Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ARID5A
Z-value: 0.81
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.17 | ARID5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg38_v1_chr2_+_96537254_96537345 | 0.04 | 5.9e-01 | Click! |
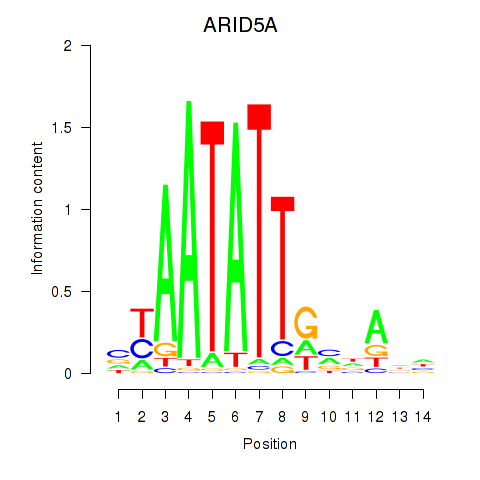
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 2.4 | 7.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 2.2 | 8.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 2.0 | 25.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.9 | 5.8 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.8 | 5.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.7 | 5.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.7 | 5.1 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 1.6 | 27.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.6 | 6.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.5 | 5.9 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 1.4 | 4.3 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.4 | 5.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.4 | 4.2 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 1.3 | 7.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.3 | 3.9 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 1.2 | 3.5 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.2 | 3.5 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 1.1 | 7.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.0 | 12.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.9 | 2.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.9 | 5.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.9 | 2.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.9 | 6.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.9 | 3.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.8 | 2.4 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.8 | 3.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.8 | 3.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.8 | 2.3 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.7 | 2.8 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.7 | 10.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.7 | 2.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.6 | 5.0 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.6 | 2.5 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.6 | 2.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 1.8 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.6 | 3.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 1.8 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.6 | 5.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.6 | 7.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.6 | 2.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.5 | 6.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.5 | 4.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 3.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.5 | 6.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 3.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 8.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 1.5 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) dehydroascorbic acid transport(GO:0070837) |
| 0.5 | 2.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.5 | 1.9 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.5 | 1.4 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.5 | 0.9 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.5 | 1.8 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 1.2 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 2.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 | 1.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 | 1.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 4.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 3.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 5.4 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.3 | 1.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 0.9 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.3 | 2.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 4.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.3 | 0.8 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.3 | 1.7 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 | 2.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 2.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 3.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 1.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.3 | 3.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 0.7 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.2 | 0.7 | GO:0060003 | copper ion export(GO:0060003) |
| 0.2 | 4.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 0.7 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 1.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.9 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 7.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 0.7 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 1.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 7.8 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 0.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 1.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 1.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 3.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 3.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 1.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 2.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.5 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) B cell selection(GO:0002339) B cell cytokine production(GO:0002368) |
| 0.2 | 1.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 1.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 1.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 1.8 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 1.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 2.6 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.7 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 13.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 2.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 2.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 3.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 3.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 3.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.5 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 3.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 1.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 2.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 2.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.6 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 3.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 1.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 3.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 3.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 3.5 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 5.4 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 1.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.2 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 11.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.0 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 3.1 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) |
| 0.1 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 3.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 4.1 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 5.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 2.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 2.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 6.2 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.1 | 0.3 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 4.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 3.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.8 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.7 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 3.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 2.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 3.4 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 5.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 3.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 2.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 3.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.4 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 3.1 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.2 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.0 | 7.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 2.0 | 25.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.5 | 4.6 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 1.4 | 4.3 | GO:0071756 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 1.2 | 3.5 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.1 | 7.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.7 | 4.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 7.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.6 | 3.7 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 3.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 6.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 3.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.4 | 1.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 2.9 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.4 | 2.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 2.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 3.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 4.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 3.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 6.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 3.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 3.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.5 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.2 | 3.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 3.0 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 1.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 3.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 2.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 12.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) postsynaptic density membrane(GO:0098839) |
| 0.1 | 2.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 2.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 2.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 33.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 1.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 2.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 7.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 2.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 9.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 6.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 11.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 7.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 4.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 11.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 3.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0044438 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 64.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 4.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 8.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 74.9 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.9 | 5.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 1.6 | 6.4 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.3 | 3.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.3 | 7.8 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 1.2 | 3.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.2 | 3.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.1 | 3.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.0 | 4.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.9 | 4.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.9 | 2.7 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.9 | 4.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.9 | 3.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.8 | 6.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.8 | 3.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.7 | 3.7 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.7 | 2.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 5.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.6 | 1.7 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.5 | 2.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.5 | 2.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 5.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 5.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 2.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.5 | 1.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.5 | 1.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.5 | 6.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.5 | 27.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.4 | 7.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 1.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 3.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.4 | 3.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 1.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 2.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.3 | 5.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 3.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 3.8 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.3 | 4.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 25.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 0.9 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.3 | 4.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 3.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 3.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.3 | 3.9 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.3 | 10.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 6.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 0.7 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 5.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 5.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 1.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 4.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.9 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 2.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 2.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 1.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 3.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 9.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 0.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 2.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 1.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 14.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.5 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 11.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 3.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.2 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 3.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 6.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 2.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 5.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 4.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 3.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 5.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 4.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 3.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 2.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.6 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.5 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 7.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 13.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 2.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 3.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 4.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.6 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 9.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 29.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 8.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 13.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 26.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 7.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 8.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 20.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 29.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 5.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 14.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 3.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.9 | 25.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.9 | 10.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 27.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.7 | 6.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 7.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 3.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 6.4 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 2.5 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 3.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 5.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 8.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 5.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 7.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 1.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 3.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 5.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 8.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 6.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 8.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 4.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 5.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 6.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 11.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 5.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 4.5 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |