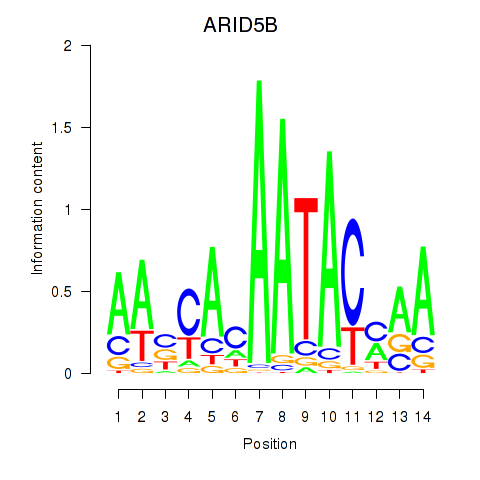
|
chr3_-_98522869
Show fit
|
7.61 |
ENST00000502288.5
ENST00000512147.5
ENST00000341181.11
ENST00000510541.5
ENST00000503621.5
ENST00000511081.5
ENST00000507874.5
ENST00000502299.5
ENST00000508659.5
ENST00000510545.5
ENST00000511667.5
ENST00000394185.6
|
CLDND1
|
claudin domain containing 1
|
|
chr3_-_98522514
Show fit
|
7.34 |
ENST00000503004.5
ENST00000506575.1
ENST00000513452.5
ENST00000515620.5
|
CLDND1
|
claudin domain containing 1
|
|
chr11_-_111923722
Show fit
|
6.81 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B
|
|
chr12_+_78863962
Show fit
|
5.61 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1
|
|
chr12_+_78864768
Show fit
|
5.26 |
ENST00000261205.9
ENST00000457153.6
|
SYT1
|
synaptotagmin 1
|
|
chr4_+_87975829
Show fit
|
3.83 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_+_84181630
Show fit
|
3.43 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta
|
|
chr3_-_79767987
Show fit
|
3.36 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1
|
|
chr4_+_76074701
Show fit
|
3.13 |
ENST00000355810.9
ENST00000349321.7
|
ART3
|
ADP-ribosyltransferase 3 (inactive)
|
|
chr12_-_91146195
Show fit
|
3.01 |
ENST00000548218.1
|
DCN
|
decorin
|
|
chr22_-_21735744
Show fit
|
2.93 |
ENST00000403503.1
|
YPEL1
|
yippee like 1
|
|
chr15_+_24823625
Show fit
|
2.92 |
ENST00000400100.5
ENST00000645002.1
ENST00000642807.1
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr7_-_24757926
Show fit
|
2.69 |
ENST00000342947.9
ENST00000419307.6
|
GSDME
|
gasdermin E
|
|
chr2_+_222861005
Show fit
|
2.67 |
ENST00000680147.1
ENST00000681009.1
ENST00000679514.1
ENST00000357430.8
|
ACSL3
|
acyl-CoA synthetase long chain family member 3
|
|
chr22_-_21735776
Show fit
|
2.65 |
ENST00000339468.8
|
YPEL1
|
yippee like 1
|
|
chr18_-_28036585
Show fit
|
2.62 |
ENST00000399380.7
|
CDH2
|
cadherin 2
|
|
chr2_+_222860942
Show fit
|
2.54 |
ENST00000392066.7
ENST00000680251.1
ENST00000679541.1
ENST00000679545.1
ENST00000680395.1
|
ACSL3
|
acyl-CoA synthetase long chain family member 3
|
|
chr17_+_3475959
Show fit
|
2.46 |
ENST00000263080.3
|
ASPA
|
aspartoacylase
|
|
chr2_+_222861059
Show fit
|
2.44 |
ENST00000681697.1
ENST00000680921.1
|
ACSL3
|
acyl-CoA synthetase long chain family member 3
|
|
chr2_+_227325225
Show fit
|
2.44 |
ENST00000353339.7
|
MFF
|
mitochondrial fission factor
|
|
chr5_+_161850597
Show fit
|
2.39 |
ENST00000634335.1
ENST00000635880.1
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1
|
|
chr4_-_86101922
Show fit
|
2.29 |
ENST00000472236.5
ENST00000641881.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr10_+_124461800
Show fit
|
2.25 |
ENST00000368842.10
ENST00000392757.8
ENST00000368839.1
|
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase
|
|
chr7_+_154052373
Show fit
|
2.20 |
ENST00000377770.8
ENST00000406326.5
|
DPP6
|
dipeptidyl peptidase like 6
|
|
chr18_+_34976928
Show fit
|
2.16 |
ENST00000591734.5
ENST00000413393.5
ENST00000589180.5
ENST00000587359.5
|
MAPRE2
|
microtubule associated protein RP/EB family member 2
|
|
chr16_-_3372666
Show fit
|
1.99 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4
|
|
chr1_+_151766655
Show fit
|
1.92 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr17_-_10549694
Show fit
|
1.88 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2
|
|
chr2_-_2324642
Show fit
|
1.82 |
ENST00000650485.1
ENST00000649207.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr12_-_8662703
Show fit
|
1.70 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr5_-_88883701
Show fit
|
1.69 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_-_74392025
Show fit
|
1.68 |
ENST00000440727.1
ENST00000409240.5
|
DCTN1
|
dynactin subunit 1
|
|
chr13_+_51861963
Show fit
|
1.62 |
ENST00000242819.7
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr12_-_8662619
Show fit
|
1.59 |
ENST00000544889.1
ENST00000543369.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr18_-_55635948
Show fit
|
1.56 |
ENST00000565124.4
ENST00000398339.5
|
TCF4
|
transcription factor 4
|
|
chr17_-_68955332
Show fit
|
1.55 |
ENST00000269080.6
ENST00000615593.4
ENST00000586539.6
ENST00000430352.6
|
ABCA8
|
ATP binding cassette subfamily A member 8
|
|
chr12_-_119803383
Show fit
|
1.49 |
ENST00000392520.2
ENST00000678677.1
ENST00000679249.1
ENST00000676849.1
|
CIT
|
citron rho-interacting serine/threonine kinase
|
|
chr12_-_8662073
Show fit
|
1.48 |
ENST00000535411.5
ENST00000540087.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr4_+_105710809
Show fit
|
1.48 |
ENST00000360505.9
ENST00000510865.5
ENST00000509336.5
|
GSTCD
|
glutathione S-transferase C-terminal domain containing
|
|
chr19_+_17215332
Show fit
|
1.47 |
ENST00000263897.10
|
USE1
|
unconventional SNARE in the ER 1
|
|
chr9_+_111525148
Show fit
|
1.43 |
ENST00000358151.8
ENST00000309235.6
ENST00000355824.7
ENST00000374374.3
|
ZNF483
|
zinc finger protein 483
|
|
chr11_-_95910665
Show fit
|
1.36 |
ENST00000674610.1
ENST00000675660.1
|
MTMR2
|
myotubularin related protein 2
|
|
chrX_-_141242512
Show fit
|
1.35 |
ENST00000358993.3
|
SPANXC
|
SPANX family member C
|
|
chr17_-_10549612
Show fit
|
1.33 |
ENST00000532183.6
ENST00000397183.6
ENST00000420805.1
|
MYH2
|
myosin heavy chain 2
|
|
chr17_+_55266216
Show fit
|
1.31 |
ENST00000573945.5
|
HLF
|
HLF transcription factor, PAR bZIP family member
|
|
chr6_-_119149124
Show fit
|
1.30 |
ENST00000368475.8
|
FAM184A
|
family with sequence similarity 184 member A
|
|
chr1_-_75611083
Show fit
|
1.27 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44 member 5
|
|
chr1_+_75724431
Show fit
|
1.26 |
ENST00000541113.6
ENST00000680805.1
|
ACADM
|
acyl-CoA dehydrogenase medium chain
|
|
chr16_+_58392462
Show fit
|
1.25 |
ENST00000318129.6
|
GINS3
|
GINS complex subunit 3
|
|
chr2_+_69013379
Show fit
|
1.17 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr1_-_168137430
Show fit
|
1.14 |
ENST00000546300.5
ENST00000271357.9
ENST00000367835.1
ENST00000537209.5
|
GPR161
|
G protein-coupled receptor 161
|
|
chr11_-_33753394
Show fit
|
1.13 |
ENST00000532057.5
ENST00000531080.5
|
FBXO3
|
F-box protein 3
|
|
chr1_+_77779763
Show fit
|
1.13 |
ENST00000646892.1
ENST00000645526.1
|
MIGA1
|
mitoguardin 1
|
|
chr10_-_73591330
Show fit
|
1.11 |
ENST00000451492.5
ENST00000681793.1
ENST00000680396.1
ENST00000413442.5
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr17_-_10549652
Show fit
|
1.09 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2
|
|
chr20_+_36091409
Show fit
|
1.09 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr2_+_69013414
Show fit
|
1.03 |
ENST00000681816.1
ENST00000482235.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr7_-_105269007
Show fit
|
1.02 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2
|
|
chr20_-_9839026
Show fit
|
1.01 |
ENST00000378429.3
ENST00000353224.10
|
PAK5
|
p21 (RAC1) activated kinase 5
|
|
chr16_+_58392391
Show fit
|
0.97 |
ENST00000426538.6
ENST00000328514.11
|
GINS3
|
GINS complex subunit 3
|
|
chr5_+_132658165
Show fit
|
0.97 |
ENST00000617259.2
ENST00000304506.7
|
IL13
|
interleukin 13
|
|
chr5_+_137889469
Show fit
|
0.97 |
ENST00000290431.5
|
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel
|
|
chr13_-_41132728
Show fit
|
0.96 |
ENST00000379485.2
|
KBTBD6
|
kelch repeat and BTB domain containing 6
|
|
chr5_+_155013755
Show fit
|
0.93 |
ENST00000435029.6
|
KIF4B
|
kinesin family member 4B
|
|
chr12_+_120496075
Show fit
|
0.92 |
ENST00000548214.1
ENST00000392508.2
|
DYNLL1
|
dynein light chain LC8-type 1
|
|
chr14_+_73097027
Show fit
|
0.92 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25
|
|
chr1_+_43389889
Show fit
|
0.92 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr12_-_8650529
Show fit
|
0.91 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr1_-_43389768
Show fit
|
0.91 |
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
|
MED8
|
mediator complex subunit 8
|
|
chr1_+_159005953
Show fit
|
0.91 |
ENST00000426592.6
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr19_-_56314788
Show fit
|
0.87 |
ENST00000592509.5
ENST00000592679.5
ENST00000683990.1
ENST00000588442.5
ENST00000593106.5
ENST00000587492.5
|
ZSCAN5A
|
zinc finger and SCAN domain containing 5A
|
|
chr12_+_120496101
Show fit
|
0.87 |
ENST00000550178.1
ENST00000550845.5
ENST00000549989.1
ENST00000552870.1
ENST00000242577.11
|
DYNLL1
|
dynein light chain LC8-type 1
|
|
chr12_-_118359105
Show fit
|
0.87 |
ENST00000541186.5
ENST00000539872.5
|
TAOK3
|
TAO kinase 3
|
|
chr1_+_77779618
Show fit
|
0.86 |
ENST00000370791.7
ENST00000443751.3
ENST00000645756.1
ENST00000643390.1
ENST00000642959.1
|
MIGA1
|
mitoguardin 1
|
|
chr12_-_8662808
Show fit
|
0.84 |
ENST00000359478.7
ENST00000396549.6
|
MFAP5
|
microfibril associated protein 5
|
|
chr22_+_19479826
Show fit
|
0.83 |
ENST00000437685.6
ENST00000263201.7
ENST00000404724.7
|
CDC45
|
cell division cycle 45
|
|
chrX_-_33211540
Show fit
|
0.83 |
ENST00000357033.9
|
DMD
|
dystrophin
|
|
chr11_+_74171266
Show fit
|
0.83 |
ENST00000328257.13
ENST00000398427.6
|
PPME1
|
protein phosphatase methylesterase 1
|
|
chrX_+_141002583
Show fit
|
0.83 |
ENST00000449283.2
|
SPANXB1
|
SPANX family member B1
|
|
chr11_-_74467531
Show fit
|
0.82 |
ENST00000310128.9
ENST00000531854.5
ENST00000526855.1
ENST00000529425.5
|
KCNE3
|
potassium voltage-gated channel subfamily E regulatory subunit 3
|
|
chr8_-_100309904
Show fit
|
0.81 |
ENST00000523481.5
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chrX_-_141698739
Show fit
|
0.78 |
ENST00000370515.3
|
SPANXD
|
SPANX family member D
|
|
chr8_-_100309368
Show fit
|
0.78 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chr1_+_186828941
Show fit
|
0.78 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA
|
|
chr14_+_64214136
Show fit
|
0.71 |
ENST00000557084.1
ENST00000458046.6
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2
|
|
chr7_+_117014881
Show fit
|
0.71 |
ENST00000422922.5
ENST00000432298.5
|
ST7
|
suppression of tumorigenicity 7
|
|
chr7_-_135510074
Show fit
|
0.69 |
ENST00000428680.6
ENST00000423368.6
ENST00000451834.5
ENST00000361528.8
ENST00000541284.5
ENST00000315544.6
|
CNOT4
|
CCR4-NOT transcription complex subunit 4
|
|
chr3_-_197297523
Show fit
|
0.69 |
ENST00000434148.1
ENST00000412364.2
ENST00000661013.1
ENST00000666007.1
ENST00000422288.6
ENST00000456699.6
ENST00000392380.6
ENST00000670935.1
ENST00000656087.1
ENST00000436682.6
ENST00000662727.1
ENST00000670455.1
ENST00000659221.1
ENST00000671185.1
ENST00000669565.1
ENST00000660898.1
ENST00000667971.1
ENST00000661453.1
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr10_+_103245887
Show fit
|
0.68 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1
|
|
chr7_-_56051544
Show fit
|
0.67 |
ENST00000395471.7
|
PSPH
|
phosphoserine phosphatase
|
|
chr1_+_148889403
Show fit
|
0.65 |
ENST00000464103.5
ENST00000534536.5
ENST00000369356.8
ENST00000369354.7
ENST00000369347.8
ENST00000369349.7
ENST00000369351.7
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr15_-_79971164
Show fit
|
0.64 |
ENST00000335661.6
ENST00000267953.4
ENST00000677151.1
|
BCL2A1
|
BCL2 related protein A1
|
|
chr9_-_32552553
Show fit
|
0.64 |
ENST00000379858.1
ENST00000360538.7
ENST00000681750.1
ENST00000680198.1
|
TOPORS
ENSG00000288684.1
|
TOP1 binding arginine/serine rich protein, E3 ubiquitin ligase
novel protein
|
|
chr2_+_205682491
Show fit
|
0.64 |
ENST00000360409.7
ENST00000450507.5
ENST00000357785.10
ENST00000417189.5
|
NRP2
|
neuropilin 2
|
|
chr1_+_209827964
Show fit
|
0.61 |
ENST00000491415.7
|
UTP25
|
UTP25 small subunit processor component
|
|
chr8_-_100309926
Show fit
|
0.60 |
ENST00000341084.7
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chr22_+_19479457
Show fit
|
0.59 |
ENST00000407835.6
ENST00000455750.6
|
CDC45
|
cell division cycle 45
|
|
chr11_-_74170792
Show fit
|
0.59 |
ENST00000680231.1
ENST00000681143.1
ENST00000679906.1
ENST00000681310.1
ENST00000414160.7
ENST00000538361.2
|
C2CD3
|
C2 domain containing 3 centriole elongation regulator
|
|
chr1_-_85276467
Show fit
|
0.57 |
ENST00000648566.1
|
BCL10
|
BCL10 immune signaling adaptor
|
|
chr6_+_24774925
Show fit
|
0.57 |
ENST00000356509.7
ENST00000230056.8
|
GMNN
|
geminin DNA replication inhibitor
|
|
chr4_-_158173042
Show fit
|
0.56 |
ENST00000592057.1
ENST00000393807.9
|
GASK1B
|
golgi associated kinase 1B
|
|
chr14_+_73569266
Show fit
|
0.55 |
ENST00000613168.1
|
ACOT2
|
acyl-CoA thioesterase 2
|
|
chr5_+_93583212
Show fit
|
0.55 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1
|
|
chr1_+_43969970
Show fit
|
0.54 |
ENST00000255108.8
ENST00000396758.6
|
DPH2
|
diphthamide biosynthesis 2
|
|
chr3_+_112086335
Show fit
|
0.54 |
ENST00000431717.6
ENST00000480282.5
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr14_+_32329256
Show fit
|
0.49 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6
|
|
chr11_+_1099730
Show fit
|
0.48 |
ENST00000674892.1
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming
|
|
chr12_-_14885845
Show fit
|
0.46 |
ENST00000539261.6
ENST00000228938.5
|
MGP
|
matrix Gla protein
|
|
chr19_+_55947832
Show fit
|
0.45 |
ENST00000291971.7
ENST00000590542.1
|
NLRP8
|
NLR family pyrin domain containing 8
|
|
chr12_+_56752449
Show fit
|
0.43 |
ENST00000554643.5
ENST00000556650.5
ENST00000554150.5
ENST00000554155.1
|
HSD17B6
|
hydroxysteroid 17-beta dehydrogenase 6
|
|
chr2_+_151409878
Show fit
|
0.42 |
ENST00000453091.6
ENST00000428287.6
ENST00000444746.7
ENST00000243326.9
ENST00000414861.6
|
RIF1
|
replication timing regulatory factor 1
|
|
chr3_+_112086364
Show fit
|
0.42 |
ENST00000264848.10
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr11_-_74170975
Show fit
|
0.41 |
ENST00000539061.6
ENST00000680645.1
ENST00000334126.12
ENST00000680718.1
|
C2CD3
|
C2 domain containing 3 centriole elongation regulator
|
|
chr4_-_158173004
Show fit
|
0.40 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B
|
|
chr6_-_30113086
Show fit
|
0.39 |
ENST00000376734.4
|
TRIM31
|
tripartite motif containing 31
|
|
chr3_+_196867856
Show fit
|
0.38 |
ENST00000445299.6
ENST00000323460.10
ENST00000419026.5
|
SENP5
|
SUMO specific peptidase 5
|
|
chr1_-_168729187
Show fit
|
0.38 |
ENST00000367817.4
|
DPT
|
dermatopontin
|
|
chr3_+_158110363
Show fit
|
0.38 |
ENST00000683137.1
|
RSRC1
|
arginine and serine rich coiled-coil 1
|
|
chr6_+_29587455
Show fit
|
0.37 |
ENST00000383640.4
|
OR2H2
|
olfactory receptor family 2 subfamily H member 2
|
|
chr4_+_94455245
Show fit
|
0.37 |
ENST00000508216.5
ENST00000514743.5
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr19_+_35358460
Show fit
|
0.35 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3
|
|
chr2_-_127675459
Show fit
|
0.34 |
ENST00000355119.9
|
LIMS2
|
LIM zinc finger domain containing 2
|
|
chr2_-_177888728
Show fit
|
0.33 |
ENST00000389683.7
|
PDE11A
|
phosphodiesterase 11A
|
|
chr2_-_169573766
Show fit
|
0.32 |
ENST00000438035.5
ENST00000453929.6
|
FASTKD1
|
FAST kinase domains 1
|
|
chr3_+_158110052
Show fit
|
0.32 |
ENST00000295930.7
ENST00000471994.5
ENST00000482822.3
ENST00000476899.6
ENST00000683899.1
ENST00000684604.1
ENST00000682164.1
ENST00000464171.5
ENST00000611884.5
ENST00000312179.10
ENST00000475278.6
|
RSRC1
|
arginine and serine rich coiled-coil 1
|
|
chr12_+_55362975
Show fit
|
0.31 |
ENST00000641576.1
|
OR6C75
|
olfactory receptor family 6 subfamily C member 75
|
|
chr11_-_66336396
Show fit
|
0.29 |
ENST00000627248.1
ENST00000311320.9
|
RIN1
|
Ras and Rab interactor 1
|
|
chr7_+_149838365
Show fit
|
0.28 |
ENST00000460379.1
ENST00000223210.5
|
ZNF862
|
zinc finger protein 862
|
|
chr1_-_113871665
Show fit
|
0.27 |
ENST00000528414.5
ENST00000460620.5
ENST00000359785.10
ENST00000420377.6
ENST00000525799.1
ENST00000538253.5
|
PTPN22
|
protein tyrosine phosphatase non-receptor type 22
|
|
chr4_+_77605807
Show fit
|
0.26 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13
|
|
chr5_+_175861628
Show fit
|
0.24 |
ENST00000509837.5
|
CPLX2
|
complexin 2
|
|
chr4_-_82848843
Show fit
|
0.23 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A, COPII coat complex component
|
|
chr5_+_162067858
Show fit
|
0.22 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr6_+_25754699
Show fit
|
0.22 |
ENST00000439485.6
ENST00000377905.9
|
SLC17A4
|
solute carrier family 17 member 4
|
|
chrX_-_11290478
Show fit
|
0.21 |
ENST00000380717.7
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr8_-_142875797
Show fit
|
0.21 |
ENST00000519285.5
|
CYP11B1
|
cytochrome P450 family 11 subfamily B member 1
|
|
chr1_+_86993009
Show fit
|
0.20 |
ENST00000370548.3
|
ENSG00000267561.2
|
novel protein
|
|
chr1_+_81306096
Show fit
|
0.18 |
ENST00000370721.5
ENST00000370727.5
ENST00000370725.5
ENST00000370723.5
ENST00000370728.5
ENST00000370730.5
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr11_+_4704782
Show fit
|
0.18 |
ENST00000380390.6
|
MMP26
|
matrix metallopeptidase 26
|
|
chr2_-_169573856
Show fit
|
0.17 |
ENST00000453153.7
ENST00000445210.1
|
FASTKD1
|
FAST kinase domains 1
|
|
chr12_-_68159732
Show fit
|
0.17 |
ENST00000229135.4
|
IFNG
|
interferon gamma
|
|
chr5_+_137889437
Show fit
|
0.16 |
ENST00000508638.5
ENST00000508883.6
ENST00000502810.5
|
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel
|
|
chr10_-_29634964
Show fit
|
0.14 |
ENST00000375398.6
ENST00000355867.8
|
SVIL
|
supervillin
|
|
chr5_+_156326735
Show fit
|
0.13 |
ENST00000435422.7
|
SGCD
|
sarcoglycan delta
|
|
chrX_-_130903187
Show fit
|
0.10 |
ENST00000432489.5
ENST00000394363.6
ENST00000338144.8
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2
|
|
chr12_+_851415
Show fit
|
0.10 |
ENST00000675631.1
|
WNK1
|
WNK lysine deficient protein kinase 1
|
|
chr6_+_30163188
Show fit
|
0.08 |
ENST00000619857.4
|
TRIM15
|
tripartite motif containing 15
|
|
chrX_-_130903224
Show fit
|
0.06 |
ENST00000370935.5
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2
|
|
chr19_-_43198079
Show fit
|
0.05 |
ENST00000597374.5
ENST00000599371.1
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4
|
|
chr8_-_132675533
Show fit
|
0.04 |
ENST00000620350.5
ENST00000518642.5
ENST00000250173.5
|
LRRC6
|
leucine rich repeat containing 6
|
|
chr2_-_218659592
Show fit
|
0.04 |
ENST00000411696.7
|
ZNF142
|
zinc finger protein 142
|
|
chr12_-_66803980
Show fit
|
0.03 |
ENST00000539540.5
ENST00000540433.5
ENST00000541947.1
ENST00000538373.1
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr12_-_56752311
Show fit
|
0.03 |
ENST00000338193.11
ENST00000550770.1
|
PRIM1
|
DNA primase subunit 1
|
|
chr13_+_102656933
Show fit
|
0.02 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2
|
|
chr2_+_89959979
Show fit
|
0.01 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28
|