Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
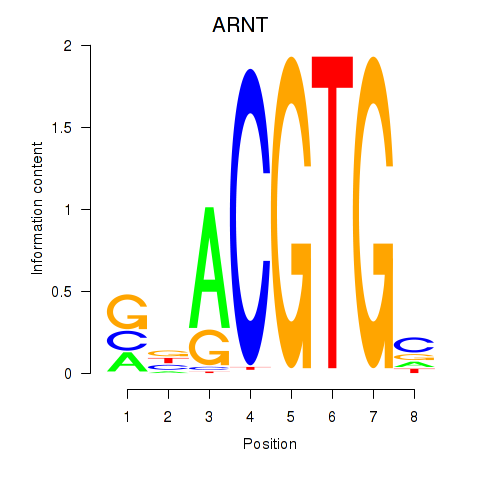
Results for ARNT
Z-value: 2.09
Transcription factors associated with ARNT
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARNT
|
ENSG00000143437.21 | ARNT |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARNT | hg38_v1_chr1_-_150876571_150876609 | -0.40 | 6.0e-10 | Click! |
Activity profile of ARNT motif
Sorted Z-values of ARNT motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARNT
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.1 | 253.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 21.6 | 107.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 17.7 | 17.7 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 17.5 | 122.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) negative regulation of megakaryocyte differentiation(GO:0045653) |
| 16.6 | 66.5 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 16.0 | 48.0 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 15.3 | 46.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 14.5 | 72.6 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 13.9 | 55.7 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 13.8 | 55.1 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 13.0 | 39.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 12.9 | 51.6 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 11.2 | 134.9 | GO:1905098 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 9.1 | 64.0 | GO:0071279 | negative regulation of mitochondrial fusion(GO:0010637) cellular response to cobalt ion(GO:0071279) |
| 8.9 | 35.6 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 8.9 | 26.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 8.2 | 24.6 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 8.0 | 32.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 7.8 | 39.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 7.8 | 31.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 7.7 | 30.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 7.7 | 23.1 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 7.7 | 30.7 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 7.6 | 38.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 7.5 | 60.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 7.5 | 119.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 7.4 | 14.8 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 7.1 | 35.6 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 7.0 | 21.0 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 7.0 | 28.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 6.8 | 34.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 6.8 | 20.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 6.7 | 20.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 6.7 | 33.5 | GO:0015862 | uridine transport(GO:0015862) |
| 6.6 | 19.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 6.5 | 19.4 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 6.2 | 18.5 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 6.1 | 18.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 6.0 | 24.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 6.0 | 113.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 6.0 | 119.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 5.8 | 17.5 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 5.7 | 17.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 5.6 | 16.9 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 5.6 | 28.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 5.5 | 33.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 5.5 | 16.5 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 5.3 | 37.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 5.1 | 46.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 4.8 | 19.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 4.8 | 19.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 4.8 | 19.1 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 4.7 | 60.7 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 4.7 | 14.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 4.6 | 13.8 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 4.5 | 40.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 4.4 | 17.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 4.3 | 21.4 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 4.2 | 20.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 4.1 | 16.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 3.9 | 11.8 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 3.8 | 49.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 3.8 | 22.7 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 3.7 | 14.9 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 3.7 | 25.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 3.7 | 40.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 3.6 | 3.6 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 3.6 | 17.8 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 3.6 | 14.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 3.5 | 10.5 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 3.5 | 14.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 3.5 | 17.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 3.3 | 16.4 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 3.2 | 12.8 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 3.2 | 19.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 3.2 | 15.8 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 3.1 | 28.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 3.1 | 9.3 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 3.0 | 53.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 2.9 | 14.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 2.9 | 26.3 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 2.8 | 19.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 2.8 | 8.5 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 2.8 | 25.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 2.8 | 14.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 2.8 | 13.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 2.7 | 56.6 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 2.7 | 26.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 2.7 | 16.0 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 2.5 | 66.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 2.5 | 22.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 2.5 | 41.7 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 2.4 | 24.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 2.3 | 20.8 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 2.2 | 45.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 2.2 | 26.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 2.2 | 8.9 | GO:0048627 | myoblast development(GO:0048627) |
| 2.1 | 44.8 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 2.0 | 12.0 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 2.0 | 31.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 2.0 | 31.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.9 | 5.7 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 1.9 | 5.7 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 1.9 | 5.6 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 1.8 | 5.5 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.7 | 12.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.7 | 6.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.7 | 15.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.6 | 11.0 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 1.6 | 3.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 1.6 | 6.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 1.5 | 19.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.5 | 7.7 | GO:0071896 | synaptic vesicle targeting(GO:0016080) protein localization to adherens junction(GO:0071896) |
| 1.5 | 14.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.5 | 5.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.5 | 20.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.5 | 50.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 1.4 | 11.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 1.4 | 127.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 1.4 | 18.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.4 | 24.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.4 | 7.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.4 | 15.6 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.4 | 8.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.3 | 99.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.3 | 22.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 1.3 | 19.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.2 | 18.6 | GO:0030656 | cobalamin transport(GO:0015889) regulation of vitamin metabolic process(GO:0030656) |
| 1.2 | 14.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 1.2 | 23.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.2 | 8.5 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.2 | 19.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.2 | 22.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.2 | 18.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 1.2 | 15.0 | GO:0045008 | depyrimidination(GO:0045008) |
| 1.1 | 5.6 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 1.1 | 7.9 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 1.1 | 97.9 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 1.1 | 5.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.1 | 20.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 1.0 | 25.7 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 1.0 | 23.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 1.0 | 2.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.0 | 8.7 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 1.0 | 7.6 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 1.0 | 4.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.0 | 3.8 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.9 | 4.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.9 | 3.6 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.9 | 12.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.9 | 23.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.9 | 33.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.9 | 6.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.8 | 3.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.8 | 5.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.8 | 29.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.7 | 12.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.7 | 38.2 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.7 | 8.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.7 | 13.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.7 | 13.4 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.7 | 26.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.7 | 57.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.6 | 21.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.6 | 47.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.6 | 3.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.6 | 23.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.6 | 4.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.6 | 6.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 9.8 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.6 | 4.0 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.6 | 24.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.6 | 13.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.5 | 6.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.5 | 4.8 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 2.6 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.5 | 3.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 33.9 | GO:0035904 | aorta development(GO:0035904) |
| 0.5 | 2.9 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 22.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.5 | 10.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.5 | 1.4 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 5.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 7.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 18.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.4 | 5.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 5.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.4 | 5.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 9.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 16.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.3 | 9.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.3 | 4.9 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 2.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) response to oleic acid(GO:0034201) |
| 0.3 | 52.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.3 | 1.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 4.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 11.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 5.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 9.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 16.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.3 | 14.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.3 | 14.2 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.3 | 4.6 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.3 | 24.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.3 | 1.8 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 6.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 6.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 10.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 0.8 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 20.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.2 | 3.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 6.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 5.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 6.7 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.2 | 4.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 5.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.2 | 10.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 0.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 15.1 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.1 | 1.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.1 | 3.6 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 0.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 15.0 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.1 | 4.8 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 | 19.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 1.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:0019344 | cysteine biosynthetic process from serine(GO:0006535) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.1 | 5.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 4.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 3.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 37.3 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 6.3 | GO:0070585 | protein localization to mitochondrion(GO:0070585) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.4 | GO:0007129 | synapsis(GO:0007129) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.7 | 253.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 12.1 | 72.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 7.7 | 122.4 | GO:0034709 | methylosome(GO:0034709) |
| 7.4 | 37.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 7.4 | 22.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 7.2 | 43.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 5.7 | 22.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 5.6 | 16.9 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 5.5 | 16.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 5.3 | 48.0 | GO:0046696 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 5.3 | 63.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 5.3 | 26.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 5.0 | 20.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 4.5 | 35.6 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 4.3 | 64.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 4.3 | 12.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 4.2 | 46.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 4.0 | 20.2 | GO:0001651 | dense fibrillar component(GO:0001651) granular component(GO:0001652) |
| 3.9 | 19.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 3.8 | 26.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 3.7 | 123.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 3.7 | 18.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 3.4 | 20.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 3.1 | 59.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 3.1 | 89.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 3.0 | 66.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 3.0 | 20.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 2.8 | 22.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 2.8 | 39.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 2.5 | 58.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 2.5 | 37.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 2.5 | 99.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 2.4 | 17.0 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 2.4 | 7.1 | GO:0044393 | microspike(GO:0044393) |
| 2.1 | 19.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 2.0 | 10.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.0 | 101.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 1.9 | 26.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.9 | 20.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.9 | 47.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.9 | 24.4 | GO:0090543 | Flemming body(GO:0090543) |
| 1.9 | 24.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.8 | 29.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.7 | 20.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.7 | 15.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 1.6 | 25.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 1.6 | 6.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.5 | 55.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.5 | 8.7 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.5 | 17.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.4 | 26.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.4 | 12.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.4 | 9.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 1.3 | 7.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 1.2 | 13.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.2 | 22.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 1.2 | 32.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 1.2 | 36.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 1.1 | 20.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.1 | 6.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 1.1 | 21.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 1.0 | 5.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 1.0 | 13.0 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 1.0 | 30.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 1.0 | 12.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 1.0 | 71.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 1.0 | 13.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.9 | 26.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.9 | 14.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.9 | 162.4 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.8 | 43.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.8 | 9.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.8 | 13.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.8 | 6.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.8 | 13.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.7 | 11.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.7 | 59.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.6 | 1.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.6 | 22.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.6 | 6.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.6 | 10.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.6 | 56.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 22.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 3.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.6 | 26.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 6.4 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 18.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.5 | 9.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.5 | 11.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.4 | 87.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 19.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 30.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 10.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 38.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 49.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.3 | 5.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 1.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 5.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.3 | 45.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 9.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 10.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 35.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.2 | 7.2 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 54.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 10.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 7.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 13.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 643.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 6.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 102.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 1.3 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.2 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 5.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 5.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 7.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 89.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 11.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 15.2 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 5.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 7.2 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 34.4 | 103.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 31.7 | 253.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 20.4 | 122.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 17.7 | 53.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 14.4 | 57.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 14.2 | 99.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 13.3 | 66.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 11.9 | 35.6 | GO:0004056 | argininosuccinate lyase activity(GO:0004056) |
| 9.5 | 38.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 9.0 | 53.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 8.6 | 43.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 8.5 | 25.6 | GO:0004639 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 8.4 | 25.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 8.3 | 41.7 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 8.1 | 40.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 8.0 | 48.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 7.7 | 30.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 7.7 | 23.1 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 7.5 | 60.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 7.0 | 28.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 7.0 | 55.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 6.7 | 20.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 6.4 | 19.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 6.2 | 24.6 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 6.1 | 30.7 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 5.6 | 16.9 | GO:0002135 | CTP binding(GO:0002135) |
| 5.6 | 33.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 5.5 | 16.5 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 5.3 | 16.0 | GO:0000035 | acyl binding(GO:0000035) |
| 5.3 | 15.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 5.1 | 20.4 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 4.7 | 51.6 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 4.6 | 32.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 4.5 | 17.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 4.3 | 29.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 4.1 | 37.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 4.0 | 35.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 3.6 | 39.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 3.5 | 14.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 3.0 | 12.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 2.9 | 66.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 2.9 | 20.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.8 | 11.3 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 2.8 | 16.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 2.7 | 11.0 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 2.7 | 10.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 2.7 | 19.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 2.6 | 39.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 2.6 | 21.1 | GO:0015288 | porin activity(GO:0015288) |
| 2.6 | 10.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 2.5 | 30.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 2.5 | 24.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 2.4 | 24.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 2.4 | 17.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 2.4 | 19.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 2.3 | 18.6 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 2.2 | 40.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 2.2 | 17.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 2.1 | 14.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 2.1 | 31.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 2.1 | 98.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 2.1 | 18.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 2.1 | 57.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 2.1 | 117.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.9 | 11.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.9 | 28.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.9 | 73.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 1.9 | 18.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.8 | 29.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.8 | 36.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 1.8 | 56.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 1.7 | 8.5 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.7 | 5.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 1.7 | 20.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 1.7 | 23.6 | GO:0031386 | protein tag(GO:0031386) |
| 1.7 | 33.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 1.6 | 21.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.6 | 9.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.5 | 21.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 1.5 | 9.0 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.4 | 46.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 1.4 | 19.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 1.4 | 5.5 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 1.3 | 10.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.3 | 12.8 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 1.2 | 12.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.2 | 22.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 1.2 | 11.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.2 | 19.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 1.2 | 3.6 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 1.2 | 3.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.2 | 8.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.2 | 24.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 1.2 | 49.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.1 | 44.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.1 | 5.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.1 | 5.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.1 | 5.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 5.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.0 | 5.2 | GO:0015093 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) ferrous iron transmembrane transporter activity(GO:0015093) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 1.0 | 16.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 1.0 | 31.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 1.0 | 15.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 1.0 | 29.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 1.0 | 2.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.9 | 26.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 3.8 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.9 | 20.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.9 | 49.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.9 | 7.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.8 | 16.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.8 | 6.7 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.8 | 14.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.8 | 60.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.8 | 5.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.8 | 7.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.8 | 19.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.8 | 2.4 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.8 | 164.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.8 | 4.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.8 | 9.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.7 | 26.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.7 | 4.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 17.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 26.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.6 | 8.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 5.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.6 | 14.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.6 | 3.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.6 | 5.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.6 | 10.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 24.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.6 | 4.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.6 | 35.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.5 | 50.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.5 | 26.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.5 | 5.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 20.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 18.8 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.5 | 28.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.4 | 11.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 14.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 6.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.4 | 9.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.4 | 12.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.4 | 3.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 14.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.4 | 34.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.4 | 4.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 17.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 4.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 3.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 5.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 6.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 46.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 14.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 9.5 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.3 | 5.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 1.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 0.8 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.3 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 9.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.3 | 7.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 13.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 4.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 8.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 5.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.8 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 10.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 59.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.2 | 4.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 2.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 6.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 12.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.5 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 6.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 7.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 165.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.1 | 10.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 6.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 7.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 6.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 2.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 3.8 | GO:0051020 | GTPase binding(GO:0051020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 134.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 4.8 | 687.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 3.4 | 171.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 2.0 | 178.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 1.4 | 20.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 1.3 | 37.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.2 | 22.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.0 | 33.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.0 | 55.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.9 | 55.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.9 | 15.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.8 | 71.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.7 | 32.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.7 | 55.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.5 | 16.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 12.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.5 | 17.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 84.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.4 | 15.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.3 | 15.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 21.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 22.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 15.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 73.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 3.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 5.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 7.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 11.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 6.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 3.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 5.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 11.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 19.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 3.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 9.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 11.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 8.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 4.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 8.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 401.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 7.0 | 105.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 5.7 | 113.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 4.8 | 43.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 3.3 | 63.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 2.6 | 93.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 2.5 | 66.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 2.5 | 98.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 2.3 | 81.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 2.2 | 71.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 2.1 | 87.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 2.1 | 18.5 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 1.8 | 53.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.6 | 29.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.6 | 46.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 1.6 | 51.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 1.5 | 34.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.5 | 87.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.4 | 39.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 1.4 | 14.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.4 | 102.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 1.3 | 32.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.3 | 33.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.3 | 36.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 1.3 | 19.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.2 | 38.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 1.2 | 87.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 1.2 | 10.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 1.1 | 22.8 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 1.1 | 30.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 1.1 | 5.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 1.1 | 12.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 1.0 | 50.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 1.0 | 49.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 1.0 | 16.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.0 | 41.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.8 | 24.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.8 | 18.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.7 | 12.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.7 | 9.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.7 | 21.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.7 | 20.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.7 | 24.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.6 | 41.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.6 | 6.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.6 | 98.5 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.6 | 19.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.6 | 13.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 5.6 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.5 | 14.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.5 | 25.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.5 | 7.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.5 | 11.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.5 | 24.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 14.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.4 | 31.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 10.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 10.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.3 | 7.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 8.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 14.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 6.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 9.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 3.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 9.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 7.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 21.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 4.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 10.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 10.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 2.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |