Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AR_NR3C2
Z-value: 1.11
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
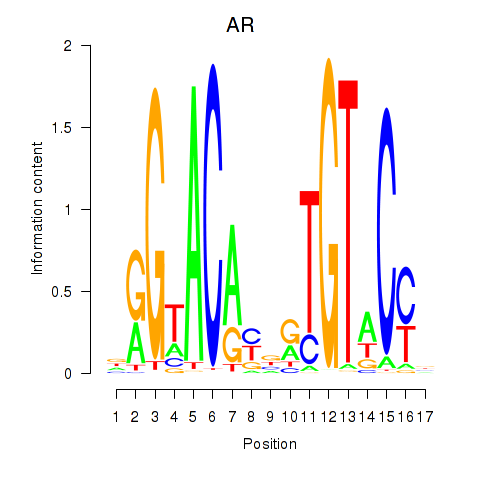
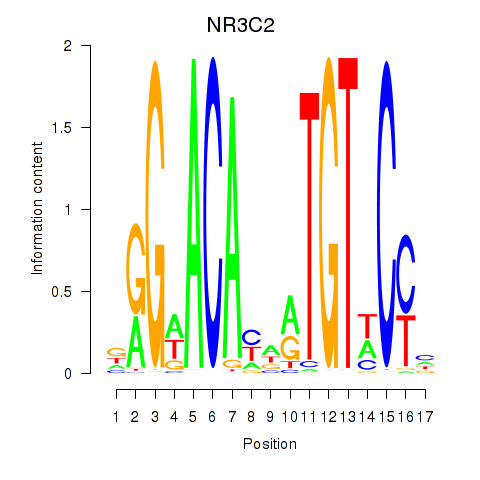
AR
|
ENSG00000169083.18 | AR |
|
NR3C2
|
ENSG00000151623.15 | NR3C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AR | hg38_v1_chrX_+_67543973_67544054 | 0.45 | 2.6e-12 | Click! |
| NR3C2 | hg38_v1_chr4_-_148442508_148442526, hg38_v1_chr4_-_148444674_148444756, hg38_v1_chr4_-_148442342_148442431 | 0.39 | 2.6e-09 | Click! |
Activity profile of AR_NR3C2 motif
Sorted Z-values of AR_NR3C2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AR_NR3C2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 49.8 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 7.9 | 55.1 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 3.7 | 80.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 1.9 | 9.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.8 | 22.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.7 | 55.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 1.4 | 4.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.8 | 6.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.7 | 5.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.7 | 9.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.7 | 2.0 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.6 | 3.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.6 | 3.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.5 | 2.0 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.5 | 1.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 6.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 53.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.4 | 3.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 1.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 3.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 2.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 1.8 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.2 | 1.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 4.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 3.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 4.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 12.8 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 4.0 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 4.0 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 3.6 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 3.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 38.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.9 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 41.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 1.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 1.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 7.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 43.0 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 4.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.6 | 54.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 5.1 | 41.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.8 | 49.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 2.5 | 22.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 3.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.5 | 3.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 2.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.3 | 4.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 2.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 4.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 143.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 2.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 4.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 51.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 38.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 5.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 3.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 27.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 5.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 25.9 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 1.7 | 5.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.8 | 49.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 3.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.5 | 3.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.5 | 3.3 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.5 | 4.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.3 | 2.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 55.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 55.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 2.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 3.1 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 9.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 1.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 123.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 9.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 4.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 7.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.7 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 4.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 3.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 5.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.5 | 9.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 56.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 25.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 6.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 22.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 12.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 10.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 54.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 8.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 23.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 9.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 4.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 4.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 6.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |