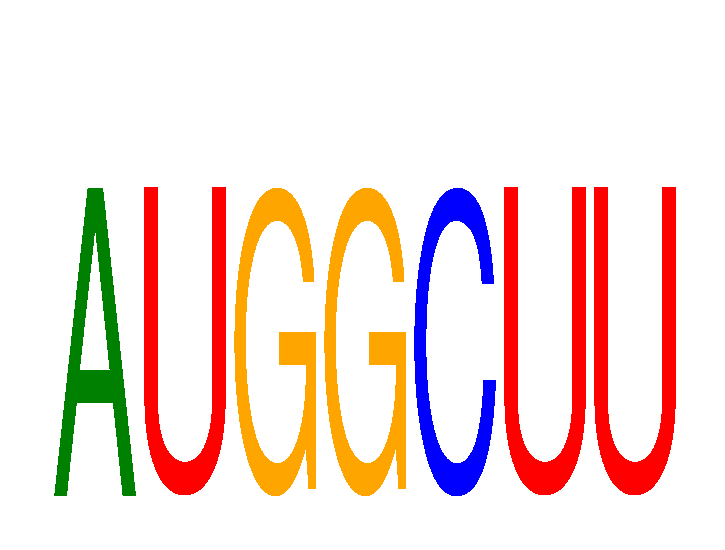Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AUGGCUU
Z-value: 0.83
miRNA associated with seed AUGGCUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-135a-5p
|
MIMAT0000428 |
|
hsa-miR-135b-5p
|
MIMAT0000758 |
Activity profile of AUGGCUU motif
Sorted Z-values of AUGGCUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AUGGCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 4.3 | 13.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 3.6 | 10.7 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 2.7 | 8.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 2.0 | 6.1 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.7 | 5.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.5 | 4.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.3 | 5.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 1.2 | 3.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.2 | 16.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.9 | 7.3 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.9 | 8.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.9 | 4.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.8 | 2.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.8 | 4.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.8 | 3.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.8 | 3.1 | GO:2000170 | positive regulation of actin filament-based movement(GO:1903116) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.7 | 3.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.7 | 2.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.7 | 2.8 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.7 | 4.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.7 | 5.9 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.6 | 7.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.6 | 1.9 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.6 | 5.0 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.6 | 3.7 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.6 | 1.8 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.6 | 7.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.6 | 2.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.6 | 2.8 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.6 | 3.9 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.5 | 4.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.5 | 3.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.5 | 7.9 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.5 | 1.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.5 | 6.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.5 | 2.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 3.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 1.9 | GO:0071878 | negative regulation of adrenergic receptor signaling pathway(GO:0071878) |
| 0.4 | 1.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.4 | 7.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.4 | 2.6 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.4 | 1.7 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.4 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 1.3 | GO:0060398 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.4 | 1.3 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.4 | 1.2 | GO:1901420 | negative regulation of prostaglandin biosynthetic process(GO:0031393) negative regulation of helicase activity(GO:0051097) negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 2.9 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 4.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 4.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.4 | 0.8 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 1.6 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.4 | 1.9 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.4 | 10.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 2.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.4 | 1.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 1.7 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 1.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 10.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.3 | 5.7 | GO:0060384 | innervation(GO:0060384) |
| 0.3 | 8.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 3.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) histone H3-K4 trimethylation(GO:0080182) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.3 | 3.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 2.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 3.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 2.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 3.0 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.3 | 1.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 15.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 2.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 3.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 0.8 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.3 | 1.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 4.6 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 | 1.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 3.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 0.5 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.2 | 0.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 3.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 2.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 2.1 | GO:0002934 | desmosome organization(GO:0002934) enamel mineralization(GO:0070166) |
| 0.2 | 2.9 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.0 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.2 | 1.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 2.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 3.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 0.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 0.8 | GO:0060611 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.2 | 1.7 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 1.1 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.2 | 0.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 8.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 3.1 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.2 | 1.9 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 3.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 1.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 3.5 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.2 | 4.3 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 0.8 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.2 | 0.3 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.2 | 8.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 11.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 1.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 5.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 1.9 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 2.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 2.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 2.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 2.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 2.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 1.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 3.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 2.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 1.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.4 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 1.8 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 3.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 3.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 1.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 1.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 4.1 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 3.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 7.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 10.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 2.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 7.8 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 1.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.9 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 6.4 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 4.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.8 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 3.0 | 8.9 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 2.6 | 13.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.7 | 5.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.5 | 7.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.4 | 20.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.3 | 4.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 1.3 | 5.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 1.0 | 3.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.9 | 17.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.7 | 3.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.6 | 3.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.6 | 7.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.6 | 3.4 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.5 | 3.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.5 | 3.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.4 | 1.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.4 | 17.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.4 | 2.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 14.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 9.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.3 | 5.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.3 | 6.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 8.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 3.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 1.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 2.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 2.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 5.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 15.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 4.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 3.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 7.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 3.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 3.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 5.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 4.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 5.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 22.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 13.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 3.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 4.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 3.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 7.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.7 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.3 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 2.2 | 8.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 2.0 | 6.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.3 | 4.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 1.2 | 10.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.1 | 4.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.9 | 3.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.9 | 4.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.8 | 2.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.7 | 4.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.6 | 13.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 9.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 12.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.6 | 1.7 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.6 | 15.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.5 | 3.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 15.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 1.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.5 | 1.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.5 | 2.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.5 | 6.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.5 | 5.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 2.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 5.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 4.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 1.3 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 1.7 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.4 | 1.2 | GO:0031696 | alpha2-adrenergic receptor activity(GO:0004938) alpha-2C adrenergic receptor binding(GO:0031696) |
| 0.4 | 1.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.4 | 11.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 3.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 15.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 2.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 1.5 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.3 | 1.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 5.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 3.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 2.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 3.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 0.8 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 1.3 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.2 | 1.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 5.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 3.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 3.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 2.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 2.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 3.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 3.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 1.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 1.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 1.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 4.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 0.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 2.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 4.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 23.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 9.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 5.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 2.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 3.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 8.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 5.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 5.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.9 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 2.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 11.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 9.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 2.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) BH3 domain binding(GO:0051434) |
| 0.0 | 2.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.0 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 4.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 10.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 16.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 12.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 4.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 4.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 20.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 10.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 6.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 14.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 8.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.3 | 4.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 12.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 4.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 8.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 12.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 4.8 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.1 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 3.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 6.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 1.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.9 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 2.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |