Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
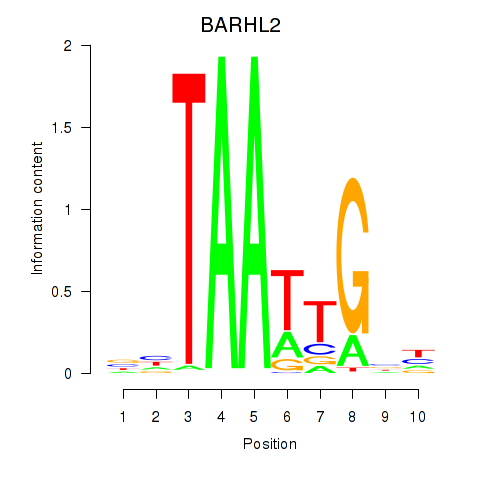
Results for BARHL2
Z-value: 0.84
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.8 | BARHL2 |
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 30.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 4.4 | 57.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 3.8 | 11.5 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) |
| 1.9 | 7.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.5 | 4.4 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.4 | 5.8 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.4 | 8.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.3 | 4.0 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.3 | 5.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 1.3 | 3.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.3 | 5.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 1.2 | 4.9 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 1.2 | 3.6 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 1.2 | 8.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 1.2 | 3.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.1 | 12.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.1 | 6.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.0 | 3.1 | GO:0009183 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.9 | 5.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.9 | 2.7 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.9 | 2.6 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.9 | 2.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.9 | 5.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.9 | 6.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.8 | 4.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.8 | 5.0 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.8 | 1.6 | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) |
| 0.8 | 4.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 11.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.8 | 7.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.8 | 5.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.7 | 3.7 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.7 | 2.2 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.7 | 5.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.7 | 8.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.7 | 6.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.7 | 2.0 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.7 | 1.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.7 | 2.0 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.6 | 5.8 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.6 | 2.5 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.6 | 4.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.6 | 3.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.6 | 6.6 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.6 | 2.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.5 | 4.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.5 | 1.6 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.5 | 1.6 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 | 9.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.5 | 2.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) |
| 0.5 | 1.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 5.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.5 | 3.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.5 | 1.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.4 | 6.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 5.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 2.6 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.4 | 3.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 9.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 5.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 5.5 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.4 | 2.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 5.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.4 | 9.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 2.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 2.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.4 | 5.6 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.3 | 2.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 | 1.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 3.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 4.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 1.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 6.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 1.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 1.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 1.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 5.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.3 | 1.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.3 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 3.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 4.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.3 | 5.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 1.6 | GO:0061517 | microglial cell activation involved in immune response(GO:0002282) macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.3 | 1.4 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.3 | 1.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.3 | 1.6 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.3 | 2.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.3 | 1.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.3 | 10.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.3 | 1.6 | GO:0050955 | thermoception(GO:0050955) |
| 0.3 | 1.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 12.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 3.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 2.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 3.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.5 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.2 | 1.4 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.2 | 0.7 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 0.9 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 2.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 3.7 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.2 | 2.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 2.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.7 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.2 | 2.0 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 3.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.2 | 19.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 5.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.9 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 1.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 2.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 9.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 1.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 3.1 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.6 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.2 | 3.9 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 1.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 1.4 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 1.8 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 2.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.6 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 0.9 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.2 | 3.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 2.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 5.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 4.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 2.6 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.2 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 1.0 | GO:1903012 | positive regulation of bone development(GO:1903012) positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 1.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 1.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 0.5 | GO:0060003 | copper ion export(GO:0060003) |
| 0.2 | 2.5 | GO:0072126 | positive regulation of glomerular mesangial cell proliferation(GO:0072126) |
| 0.2 | 2.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 5.8 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.6 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 7.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.7 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 7.9 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 2.8 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 4.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 5.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 3.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 10.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 2.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 2.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 4.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 1.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 1.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 12.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.6 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.5 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 4.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 2.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 2.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.3 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.1 | 18.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 3.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 3.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 1.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 6.9 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 6.5 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 0.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.5 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.1 | 0.3 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 1.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.2 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 3.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.9 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 2.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.9 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 2.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.4 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 3.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 4.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.0 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 3.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.3 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 3.6 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.0 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 4.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 1.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 1.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.7 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.4 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 2.6 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 1.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.0 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 1.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 2.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 1.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) |
| 0.0 | 0.3 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.9 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 2.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0035990 | tendon development(GO:0035989) tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 2.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 1.1 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 6.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.4 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 1.7 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.0 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.6 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 3.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.5 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.6 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.6 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.7 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.0 | GO:0010887 | detection of hormone stimulus(GO:0009720) detection of endogenous stimulus(GO:0009726) negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.1 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.7 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 1.3 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 81.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.9 | 11.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.1 | 8.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.3 | 5.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.8 | 3.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.7 | 2.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.7 | 5.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.7 | 4.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 3.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.6 | 1.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.6 | 7.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 11.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 6.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 3.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.5 | 6.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 4.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 3.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 2.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 7.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 2.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 3.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 2.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 5.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 3.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.4 | 5.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 13.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 4.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 6.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 5.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 33.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 3.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.3 | 1.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 1.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 6.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 5.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 1.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 3.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.9 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.2 | 4.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 12.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 8.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 7.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 8.9 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 5.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 6.1 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 3.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 16.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 2.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 2.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 7.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.0 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 0.8 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 2.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 7.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 8.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 4.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 4.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 4.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 23.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 7.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 5.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 6.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 8.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 6.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 9.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 10.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 2.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 10.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 11.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.0 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.9 | 11.5 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 2.1 | 8.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 2.1 | 6.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.4 | 1.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.3 | 4.0 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.3 | 5.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 1.3 | 7.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 1.2 | 6.9 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 1.1 | 4.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 3.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.0 | 9.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.0 | 5.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.9 | 3.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.8 | 3.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 5.6 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.8 | 3.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.8 | 3.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.7 | 2.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 6.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 2.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.6 | 5.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 51.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.6 | 1.8 | GO:0042806 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 0.6 | 3.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.6 | 1.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.5 | 7.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 2.5 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.5 | 8.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 6.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 2.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.4 | 5.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 2.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 46.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 4.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 1.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 5.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 7.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 2.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 4.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.4 | 4.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 1.2 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.4 | 4.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 7.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 8.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 2.0 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.3 | 5.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 1.3 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.3 | 1.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.3 | 1.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 4.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 5.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.1 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.3 | 3.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 5.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 2.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 3.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 1.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.2 | 1.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 1.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 5.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 3.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 4.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 6.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 1.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 1.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 4.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 6.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 1.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.9 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 1.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 3.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 5.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 0.5 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 4.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 1.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 2.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 2.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 1.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 3.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 2.0 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.9 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 3.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 2.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.7 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 9.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 3.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 4.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 4.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 13.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 3.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 9.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 4.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 6.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.9 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 3.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 2.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 2.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 12.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.7 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 7.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 4.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 7.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 6.6 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 3.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 6.3 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 17.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 2.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 5.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0033691 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) sialic acid binding(GO:0033691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 68.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.1 | 30.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 24.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 7.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 8.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 33.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 7.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 11.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 8.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 9.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 5.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 9.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 9.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 13.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 60.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.9 | 20.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 9.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 35.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 7.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.4 | 6.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 11.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.3 | 7.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.3 | 9.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 5.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 15.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 9.0 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 5.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 7.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 7.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 1.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 1.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 8.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 6.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 4.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 4.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 7.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 5.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 5.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.9 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |