Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
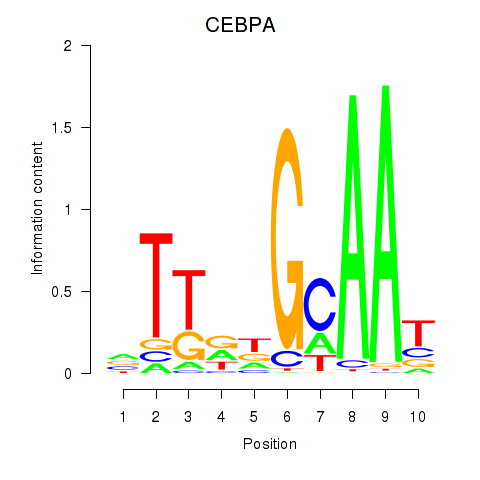
Results for CEBPA
Z-value: 1.61
Transcription factors associated with CEBPA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPA
|
ENSG00000245848.3 | CEBPA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPA | hg38_v1_chr19_-_33302524_33302542 | 0.63 | 3.9e-25 | Click! |
Activity profile of CEBPA motif
Sorted Z-values of CEBPA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.8 | 69.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 9.5 | 47.5 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 8.7 | 34.7 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 6.4 | 19.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 5.7 | 39.9 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 5.4 | 26.8 | GO:0015688 | response to mycotoxin(GO:0010046) iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 5.0 | 15.0 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 4.3 | 8.6 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 4.3 | 30.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 4.2 | 12.7 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 4.0 | 12.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 3.9 | 15.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 3.8 | 11.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 3.6 | 3.6 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 3.4 | 17.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 3.4 | 10.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 3.2 | 35.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 3.0 | 9.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 3.0 | 9.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 2.8 | 16.9 | GO:0032571 | response to vitamin K(GO:0032571) |
| 2.7 | 8.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 2.5 | 12.4 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 2.3 | 6.9 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 2.3 | 15.8 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 2.2 | 22.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 2.1 | 8.6 | GO:0002384 | hepatic immune response(GO:0002384) |
| 2.1 | 12.7 | GO:0035624 | receptor transactivation(GO:0035624) |
| 2.1 | 27.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 2.0 | 6.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 2.0 | 6.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 2.0 | 5.9 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 2.0 | 5.9 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.9 | 9.7 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 1.9 | 5.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.9 | 9.3 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 1.9 | 24.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.7 | 34.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.7 | 5.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.6 | 55.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.6 | 13.0 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.6 | 4.8 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.6 | 4.7 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 1.6 | 4.7 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 1.5 | 6.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 1.5 | 13.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.5 | 16.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.4 | 5.8 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 1.4 | 26.9 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 1.4 | 4.2 | GO:0009720 | detection of hormone stimulus(GO:0009720) response to high density lipoprotein particle(GO:0055099) |
| 1.4 | 5.6 | GO:0060620 | regulation of high-density lipoprotein particle clearance(GO:0010982) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.4 | 11.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 1.4 | 26.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.3 | 7.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.3 | 7.5 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 1.2 | 5.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.2 | 3.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 1.1 | 6.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.1 | 3.3 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 1.1 | 6.4 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 1.1 | 9.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 1.0 | 5.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 1.0 | 3.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.0 | 4.0 | GO:1903627 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 1.0 | 3.0 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 1.0 | 3.9 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 1.0 | 4.8 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 1.0 | 5.7 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.9 | 12.3 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.9 | 12.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.9 | 1.9 | GO:1904640 | response to methionine(GO:1904640) |
| 0.9 | 2.8 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.9 | 29.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.9 | 21.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.9 | 2.7 | GO:2000722 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 0.9 | 8.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.9 | 7.9 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.9 | 3.5 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.8 | 12.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.8 | 3.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.8 | 10.8 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.8 | 3.9 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.8 | 48.9 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.8 | 3.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.8 | 54.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.8 | 6.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.7 | 14.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.7 | 2.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.7 | 5.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.7 | 5.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.7 | 10.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.7 | 4.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.7 | 2.7 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.7 | 2.0 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.6 | 2.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.6 | 5.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.6 | 2.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 3.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.6 | 6.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 3.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.6 | 3.5 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.6 | 3.5 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.6 | 8.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.6 | 4.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.6 | 2.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.6 | 4.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 4.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.6 | 31.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.5 | 4.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.5 | 0.5 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.5 | 5.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.5 | 0.5 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.5 | 3.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.5 | 7.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.5 | 2.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 1.4 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.4 | 26.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.4 | 7.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.4 | 1.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.4 | 4.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 1.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 3.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 2.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.4 | 1.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 4.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.4 | 1.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 4.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 2.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.3 | 9.1 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.3 | 2.7 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.3 | 5.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.3 | 2.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 2.8 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 6.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.3 | 1.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 2.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.3 | 0.9 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.3 | 2.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.3 | 2.0 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 1.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 3.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.3 | 3.9 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 2.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 5.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 3.3 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.2 | 0.7 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 2.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 3.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 2.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 2.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.2 | 5.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 3.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 2.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 8.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 9.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 16.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 5.5 | GO:0016246 | RNA interference(GO:0016246) |
| 0.2 | 6.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 27.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 5.9 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 8.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 5.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 4.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 6.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 1.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 66.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.2 | 1.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 0.5 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 5.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 10.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 3.2 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 0.6 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.1 | 1.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 1.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 4.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 11.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 2.9 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 3.1 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 1.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 5.1 | GO:0090102 | cochlea development(GO:0090102) |
| 0.1 | 1.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.4 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 9.1 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 1.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 2.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 2.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.7 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 3.6 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.1 | 3.4 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 2.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 14.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.2 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.1 | 2.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 2.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 5.1 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 8.4 | GO:0015992 | proton transport(GO:0015992) |
| 0.1 | 0.1 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 1.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 2.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 6.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 3.2 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 1.8 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 2.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 1.0 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 2.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 2.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.9 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 3.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 1.8 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.3 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.2 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 1.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 5.3 | 47.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 4.8 | 19.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.2 | 12.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.2 | 22.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 2.1 | 8.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 2.1 | 194.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 2.0 | 26.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.0 | 25.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.0 | 5.9 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.9 | 13.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.8 | 9.1 | GO:0001652 | granular component(GO:0001652) |
| 1.8 | 34.7 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 1.7 | 8.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.6 | 10.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 1.4 | 4.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.1 | 12.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 1.0 | 11.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.0 | 10.7 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.9 | 7.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.9 | 10.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.9 | 9.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.8 | 6.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.8 | 6.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.7 | 15.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.6 | 84.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.6 | 68.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.6 | 15.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 9.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.5 | 4.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 2.0 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.5 | 2.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 5.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 3.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 3.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 6.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 65.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 3.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 2.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 2.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 2.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 1.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 4.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 15.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 1.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 2.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 3.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 2.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 7.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 30.9 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.2 | 41.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 3.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 24.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 1.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 6.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.9 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 13.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 5.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 49.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 5.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 2.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 15.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 3.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 9.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 4.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 7.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 20.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 9.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 10.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 13.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 103.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 9.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 9.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 6.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 77.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 5.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 3.1 | GO:0044215 | host(GO:0018995) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 4.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 7.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.2 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.4 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.4 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 52.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 11.5 | 69.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 7.1 | 35.5 | GO:0047374 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 4.3 | 30.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 3.8 | 26.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 3.5 | 10.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 3.5 | 10.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 3.3 | 39.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 3.1 | 12.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 2.9 | 8.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.9 | 8.6 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 2.7 | 8.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.5 | 10.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 2.5 | 10.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 2.3 | 9.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 2.2 | 6.6 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 2.2 | 26.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.1 | 10.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.0 | 6.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 2.0 | 7.8 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.9 | 7.5 | GO:0035473 | lipase binding(GO:0035473) |
| 1.8 | 7.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.7 | 3.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.7 | 26.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 1.6 | 16.1 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.5 | 7.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.5 | 5.9 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 1.5 | 56.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.4 | 36.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 1.4 | 16.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.3 | 3.9 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.1 | 5.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.1 | 3.3 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) |
| 1.0 | 4.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 1.0 | 5.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.0 | 20.8 | GO:0005522 | profilin binding(GO:0005522) |
| 1.0 | 4.0 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 1.0 | 6.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.0 | 6.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 5.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.9 | 4.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.9 | 18.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.8 | 5.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.8 | 2.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.8 | 2.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.8 | 3.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.8 | 17.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.8 | 3.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 11.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.8 | 28.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.8 | 10.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.7 | 5.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.7 | 76.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.7 | 13.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.7 | 3.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 7.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.6 | 3.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.6 | 1.8 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.6 | 3.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 26.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.6 | 8.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.5 | 2.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 11.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.5 | 29.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.5 | 3.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.5 | 3.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.5 | 4.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 5.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 1.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.5 | 6.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.5 | 2.0 | GO:0008422 | glucosylceramidase activity(GO:0004348) beta-glucosidase activity(GO:0008422) |
| 0.5 | 1.9 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.4 | 3.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 6.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 4.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 4.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 1.7 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.4 | 16.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 1.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.4 | 1.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 12.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 14.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 11.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.4 | 6.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 12.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 7.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.4 | 1.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.4 | 5.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.4 | 5.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 12.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 1.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 4.8 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.3 | 3.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.3 | 9.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 2.8 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.3 | 2.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 5.6 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.3 | 22.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 77.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 2.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 2.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 8.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 3.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 5.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 1.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 5.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 3.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 3.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 26.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 8.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 23.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.2 | 2.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 14.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 5.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 2.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 16.9 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.2 | 1.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 23.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 10.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 3.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 15.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 3.7 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 7.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 7.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 10.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 10.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 7.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 9.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 2.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.8 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 4.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 7.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 1.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 5.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 5.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 14.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 24.7 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 6.8 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 6.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 3.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 2.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0070679 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0097617 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 35.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.3 | 46.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.9 | 25.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.9 | 26.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.9 | 11.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.8 | 3.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.7 | 72.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.6 | 29.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.6 | 8.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.6 | 86.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.6 | 22.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.5 | 21.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 14.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.4 | 12.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 30.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 55.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 7.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 16.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 72.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 14.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 10.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 15.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 51.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 14.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 8.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 11.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 5.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 71.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 3.4 | 43.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 2.5 | 2.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 2.4 | 45.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 2.0 | 22.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.0 | 46.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.7 | 27.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.7 | 31.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.6 | 71.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.5 | 15.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.5 | 26.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.2 | 29.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.1 | 25.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.0 | 14.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.0 | 3.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.9 | 18.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.7 | 13.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 31.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.6 | 2.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 32.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 6.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.6 | 5.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 8.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.5 | 32.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 7.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 17.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 8.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 8.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 11.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 7.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 2.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 12.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 7.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 2.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 8.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 15.7 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.3 | 8.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 5.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 10.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 18.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 8.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 1.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 5.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 8.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 3.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 4.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 37.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 12.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 10.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 2.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 3.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 5.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 4.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.5 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 4.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 5.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 11.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 2.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |