Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for CPEB1
Z-value: 1.09
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.10 | CPEB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg38_v1_chr15_-_82647336_82647386 | -0.41 | 2.4e-10 | Click! |
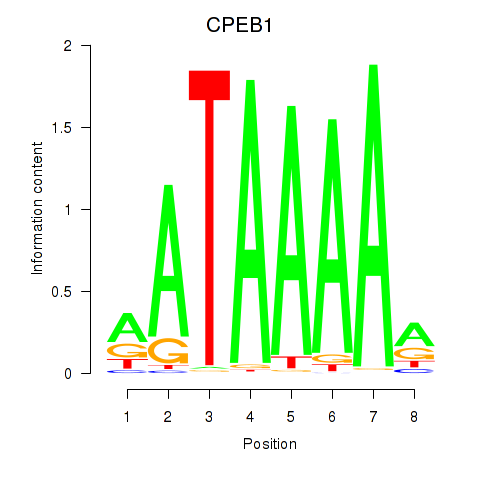
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 47.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 3.6 | 14.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 3.5 | 46.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 3.2 | 15.8 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 3.1 | 9.4 | GO:0051821 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) development involved in symbiotic interaction(GO:0044111) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 2.4 | 9.7 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 2.2 | 15.4 | GO:0008215 | spermine metabolic process(GO:0008215) spermidine biosynthetic process(GO:0008295) |
| 2.2 | 8.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 2.0 | 10.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.0 | 12.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 2.0 | 10.0 | GO:0015862 | uridine transport(GO:0015862) |
| 2.0 | 6.0 | GO:0061573 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 2.0 | 7.9 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 1.9 | 17.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.8 | 7.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.8 | 3.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 1.7 | 7.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 1.7 | 8.6 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 1.7 | 25.0 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 1.6 | 21.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.6 | 4.7 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.6 | 14.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.5 | 6.0 | GO:0060585 | rhythmic synaptic transmission(GO:0060024) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.4 | 5.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 1.4 | 5.7 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 1.4 | 10.9 | GO:0030091 | protein repair(GO:0030091) |
| 1.4 | 10.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.3 | 9.2 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 1.3 | 11.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.2 | 4.9 | GO:0061073 | ciliary body morphogenesis(GO:0061073) endocardial cushion cell differentiation(GO:0061443) |
| 1.2 | 7.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.2 | 15.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.1 | 6.8 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 1.1 | 3.3 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 1.0 | 4.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 1.0 | 5.2 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 1.0 | 8.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.0 | 11.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.9 | 6.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.9 | 2.8 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.9 | 4.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.9 | 2.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.9 | 8.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.9 | 8.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.9 | 3.5 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.9 | 3.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.9 | 36.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.8 | 6.7 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.8 | 5.8 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.8 | 2.5 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.8 | 3.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.8 | 5.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.8 | 6.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.8 | 3.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.8 | 4.5 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.7 | 8.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.7 | 3.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.7 | 5.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.7 | 11.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.7 | 8.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.7 | 5.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.7 | 3.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 27.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.6 | 20.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.6 | 6.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.6 | 1.8 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.6 | 3.5 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.6 | 3.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.6 | 7.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.6 | 7.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.6 | 4.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.5 | 3.6 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.5 | 4.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 6.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.5 | 2.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.5 | 3.5 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.5 | 2.9 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.5 | 1.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.5 | 2.3 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.5 | 6.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 4.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 4.4 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.4 | 4.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.4 | 2.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.4 | 1.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 24.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.4 | 3.9 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.4 | 5.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.4 | 1.6 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 | 6.0 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.4 | 1.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.4 | 5.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.4 | 1.5 | GO:1900450 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.4 | 2.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.4 | 10.6 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.4 | 10.1 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.4 | 7.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 1.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.3 | 3.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 6.5 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.3 | 2.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 1.6 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 2.2 | GO:0010710 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 0.3 | 12.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.3 | 2.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.3 | 2.9 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 2.7 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.3 | 4.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 1.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.7 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.2 | 9.6 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 7.1 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.2 | 5.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 5.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 4.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 10.9 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 26.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 7.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 2.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 10.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.2 | GO:2000230 | response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.5 | GO:1901624 | T cell extravasation(GO:0072683) negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.2 | 17.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 0.8 | GO:1900376 | regulation of melanin biosynthetic process(GO:0048021) regulation of secondary metabolite biosynthetic process(GO:1900376) |
| 0.2 | 5.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 2.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.8 | GO:0021886 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 7.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.6 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 1.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.3 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.1 | 1.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 19.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 6.5 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 1.9 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 1.5 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 0.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.0 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 8.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 2.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 7.0 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 4.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 25.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 2.2 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 11.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 15.8 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 4.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 6.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 3.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 4.7 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 2.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.1 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 3.4 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0070572 | positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 2.2 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 1.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 4.3 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 1.1 | GO:0034605 | cellular response to heat(GO:0034605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.0 | GO:0008623 | CHRAC(GO:0008623) |
| 5.3 | 15.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.0 | 8.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 2.9 | 17.6 | GO:1990357 | terminal web(GO:1990357) |
| 2.9 | 8.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.4 | 7.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 2.3 | 9.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 2.0 | 6.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.8 | 44.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.4 | 15.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.4 | 8.2 | GO:0002177 | manchette(GO:0002177) |
| 1.3 | 14.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.3 | 12.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 1.2 | 27.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 1.2 | 3.5 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.1 | 4.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 1.1 | 7.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.0 | 10.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.0 | 12.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.9 | 8.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.8 | 2.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.8 | 9.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.7 | 7.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 8.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.5 | 11.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 5.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.5 | 46.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 5.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 7.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 17.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 4.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 2.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.4 | 4.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 7.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 3.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 2.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.3 | 4.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 5.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.3 | 3.5 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 1.4 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 8.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 6.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 10.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.2 | 1.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 4.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 4.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 36.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 6.9 | GO:0016514 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.2 | 4.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 1.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 6.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 17.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 10.3 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 4.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 36.0 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 51.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 15.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 8.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.3 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.1 | 6.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 21.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 9.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 12.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 23.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 4.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 10.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 0.4 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 2.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 32.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 15.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 5.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 8.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 7.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 3.8 | GO:0045177 | apical part of cell(GO:0045177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 36.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 4.7 | 14.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 3.1 | 28.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 2.3 | 9.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 2.2 | 10.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 2.2 | 10.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.1 | 46.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.9 | 42.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 1.8 | 8.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.7 | 7.0 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 1.6 | 21.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.6 | 14.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.6 | 7.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.5 | 4.5 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 1.4 | 10.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.4 | 5.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 1.4 | 6.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.3 | 9.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 1.3 | 23.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.3 | 8.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.0 | 6.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.0 | 5.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.9 | 2.7 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.9 | 4.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.9 | 3.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.8 | 17.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.8 | 5.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.8 | 10.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.8 | 2.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.8 | 2.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.8 | 3.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.7 | 11.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.7 | 5.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.7 | 6.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 40.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.6 | 3.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.6 | 3.5 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.6 | 8.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 3.5 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.5 | 4.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 8.6 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.5 | 10.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.5 | 7.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 17.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 12.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 3.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 19.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.4 | 4.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.4 | 10.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 5.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 4.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 23.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.3 | 2.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.3 | 11.0 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.3 | 13.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 4.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 11.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 6.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 2.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 5.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 3.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 15.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 12.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 2.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 15.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 1.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 4.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 1.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 7.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 4.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 6.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 4.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 2.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 7.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 3.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 8.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 5.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 5.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 1.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 7.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 3.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 8.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 6.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 4.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 2.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 5.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 18.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 41.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 2.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 1.2 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 5.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 5.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 11.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 2.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 22.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 7.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 11.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 7.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 30.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 21.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 31.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.4 | 15.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 47.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.4 | 21.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 16.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 11.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 3.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 12.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 8.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 3.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 16.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 4.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 5.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 9.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 4.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 13.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 10.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 9.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 7.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 8.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 9.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 5.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 4.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 42.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 1.2 | 43.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 1.0 | 13.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.0 | 17.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.8 | 15.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.7 | 25.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 15.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 34.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 12.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 4.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 5.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 7.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.4 | 20.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 16.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 16.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 10.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 5.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 15.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 11.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.3 | 7.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 12.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 11.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 7.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 12.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 4.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 3.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 15.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 10.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 10.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 10.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 3.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 3.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 4.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 39.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 5.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 8.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 7.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 7.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 5.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 2.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 3.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 3.7 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |