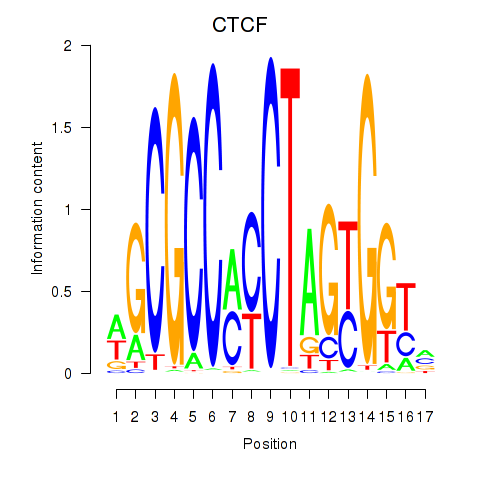
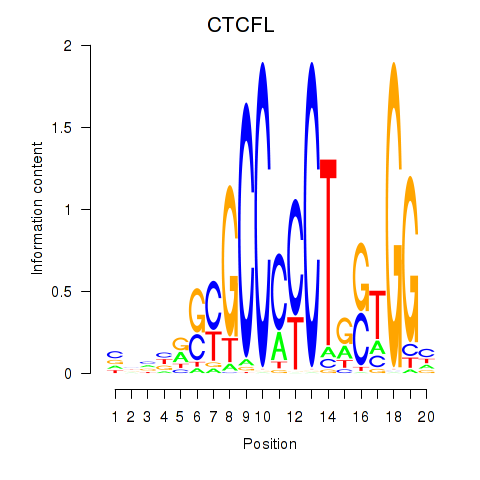
Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for CTCF_CTCFL
Z-value: 0.91
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CTCF
|
ENSG00000102974.16 | CTCF |
|
CTCFL
|
ENSG00000124092.13 | CTCFL |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCFL | hg38_v1_chr20_-_57525097_57525108 | -0.40 | 1.3e-09 | Click! |
| CTCF | hg38_v1_chr16_+_67562514_67562594 | 0.37 | 2.8e-08 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 15.5 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 5.6 | 16.7 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 5.1 | 15.4 | GO:0097069 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 4.9 | 14.6 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 4.8 | 14.4 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 4.2 | 21.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 3.9 | 34.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 3.1 | 18.8 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 3.0 | 9.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 3.0 | 12.0 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 2.7 | 16.4 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 2.7 | 5.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 2.4 | 9.7 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 2.4 | 7.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 2.4 | 9.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 2.2 | 13.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 2.2 | 6.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) mitochondrial DNA repair(GO:0043504) regulation of myofibroblast differentiation(GO:1904760) |
| 2.1 | 4.2 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 2.0 | 6.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.9 | 9.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.9 | 1.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 1.9 | 5.7 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.9 | 7.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.8 | 9.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.8 | 7.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 1.8 | 12.6 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 1.5 | 4.5 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 1.4 | 4.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 1.4 | 6.9 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.4 | 6.9 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.4 | 8.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 1.3 | 22.9 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 1.3 | 11.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.3 | 6.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 1.3 | 10.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.2 | 3.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.2 | 9.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.2 | 2.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 1.1 | 2.2 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 1.1 | 6.7 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.1 | 3.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 1.1 | 5.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.1 | 3.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 1.0 | 10.5 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 1.0 | 4.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.0 | 14.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.0 | 5.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.9 | 7.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.9 | 2.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.9 | 3.6 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.9 | 16.8 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.9 | 3.5 | GO:2000653 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.9 | 5.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.9 | 2.6 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of positive thymic T cell selection(GO:1902232) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.8 | 10.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.8 | 4.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.8 | 4.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.8 | 2.5 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.8 | 2.5 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.8 | 6.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.8 | 8.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.8 | 2.4 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.8 | 6.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.8 | 5.5 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.8 | 8.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.8 | 9.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.7 | 1.5 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.7 | 15.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.7 | 6.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.7 | 2.1 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.7 | 32.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.7 | 2.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.7 | 50.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.7 | 8.0 | GO:1905098 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.6 | 16.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 1.9 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.6 | 1.9 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.6 | 1.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.6 | 3.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.6 | 3.7 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.6 | 1.2 | GO:0002339 | B cell selection(GO:0002339) |
| 0.6 | 14.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.6 | 1.8 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.6 | 5.8 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.6 | 8.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.6 | 4.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.5 | 3.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.5 | 9.1 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.5 | 1.6 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.5 | 4.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 3.7 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.5 | 4.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.5 | 3.6 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.5 | 2.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.5 | 1.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.5 | 1.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.5 | 1.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.5 | 1.4 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.5 | 1.4 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.5 | 4.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.5 | 9.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 7.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 1.8 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.4 | 6.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.4 | 1.8 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.4 | 4.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.4 | 3.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 | 3.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.4 | 4.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.4 | 3.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 | 1.7 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.4 | 1.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) activation of meiosis(GO:0090427) |
| 0.4 | 2.4 | GO:0045963 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.4 | 32.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.4 | 1.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.4 | 1.5 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.4 | 11.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.4 | 3.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 | 4.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.4 | 1.5 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 2.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.4 | 2.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.4 | 2.9 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.4 | 2.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 | 3.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 2.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 3.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 2.4 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 1.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.3 | 1.3 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.3 | 2.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 2.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 20.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 1.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.3 | 0.6 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.3 | 3.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 13.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.3 | 10.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 3.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 4.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 2.6 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 10.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 6.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 5.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 2.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 2.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 3.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 0.8 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.5 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.2 | 2.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 1.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 10.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 1.4 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 8.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 35.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 4.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 6.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.2 | 0.9 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 4.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.2 | 3.9 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.2 | 5.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 3.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.6 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.2 | 1.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 3.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 3.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 | 1.0 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.6 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.2 | 18.6 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 1.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 6.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 6.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 2.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.9 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.2 | 14.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 2.8 | GO:1901838 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.2 | 1.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 0.7 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 7.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 1.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 0.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 2.7 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.2 | 2.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.2 | 2.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 2.7 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 3.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 2.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.6 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 6.4 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 0.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 4.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 5.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 0.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 6.7 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 2.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.9 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 8.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 5.3 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 0.6 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.0 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.1 | 0.7 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 4.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.9 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.6 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.5 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 3.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 10.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 1.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 2.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 4.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 9.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.6 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 23.9 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 8.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.1 | 10.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.3 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.3 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.5 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 0.9 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.4 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 1.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.2 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.9 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 1.6 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.3 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 2.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 5.7 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 2.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.7 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 5.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.7 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.0 | 0.5 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 1.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.8 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 51.9 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 5.1 | 15.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 4.2 | 16.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 3.1 | 15.6 | GO:0089701 | U2AF(GO:0089701) |
| 2.8 | 11.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 2.7 | 26.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 2.6 | 18.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 2.1 | 6.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 2.1 | 18.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.9 | 21.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.8 | 19.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 1.7 | 18.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 1.6 | 16.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.5 | 4.5 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 1.5 | 23.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.4 | 4.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.4 | 9.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.2 | 5.0 | GO:0071942 | XPC complex(GO:0071942) |
| 1.2 | 12.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 1.1 | 5.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.1 | 44.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.1 | 3.3 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 1.0 | 12.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.0 | 5.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.0 | 28.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.0 | 10.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.0 | 3.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 1.0 | 2.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.9 | 3.7 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.9 | 2.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.9 | 15.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.9 | 2.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.8 | 3.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.8 | 4.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.8 | 9.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.8 | 3.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.7 | 9.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.7 | 6.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.7 | 8.0 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.7 | 3.3 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.7 | 11.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.6 | 5.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.6 | 11.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 5.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 3.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 3.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 3.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.5 | 6.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 2.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.5 | 1.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 2.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.4 | 3.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 46.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 2.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 2.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.4 | 2.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 3.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 3.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 19.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 1.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.3 | 5.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 3.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 4.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 3.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 5.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 3.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 3.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 2.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 3.3 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.3 | 4.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.3 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 3.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 1.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.3 | 5.6 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.3 | 4.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 11.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 8.1 | GO:0016469 | proton-transporting two-sector ATPase complex(GO:0016469) |
| 0.2 | 1.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 0.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 1.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 1.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 16.9 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 2.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 1.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 6.4 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.2 | 25.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 3.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 17.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 3.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 13.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.2 | 5.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 11.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.0 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 3.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 0.5 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.2 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 7.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 1.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 13.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 21.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 8.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.4 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 2.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 7.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 11.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.2 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 5.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 10.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 11.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 3.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 7.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 4.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 3.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 19.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 4.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 6.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 33.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 6.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 4.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 11.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.4 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 5.1 | 15.4 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 4.9 | 14.8 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 3.0 | 18.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 2.4 | 12.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 2.1 | 21.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 2.0 | 8.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 1.9 | 5.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.8 | 5.5 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 1.8 | 9.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 1.8 | 7.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 1.7 | 15.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.7 | 6.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.7 | 5.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.6 | 8.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 1.6 | 28.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 1.5 | 4.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 1.4 | 11.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.4 | 14.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 1.4 | 5.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.4 | 18.0 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 1.4 | 9.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 12.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.3 | 6.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 1.2 | 11.2 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 1.2 | 6.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 1.2 | 7.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.2 | 10.8 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 1.2 | 5.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.2 | 5.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.1 | 16.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 1.1 | 13.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.0 | 3.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.0 | 5.9 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.0 | 9.7 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.9 | 7.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.9 | 8.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.9 | 2.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.9 | 5.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.9 | 2.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.8 | 11.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.8 | 6.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.8 | 5.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.8 | 8.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.8 | 14.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 11.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 3.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.7 | 5.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.7 | 15.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.7 | 11.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.7 | 4.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.7 | 5.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.7 | 2.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.7 | 2.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.7 | 4.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.7 | 3.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.7 | 5.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.7 | 2.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.6 | 3.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.6 | 3.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.6 | 28.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.6 | 11.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.6 | 2.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.6 | 25.3 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.6 | 9.1 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.6 | 3.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.5 | 4.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 1.6 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.5 | 31.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.5 | 2.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 20.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.5 | 15.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 7.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.5 | 4.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.5 | 6.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.4 | 3.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.4 | 4.2 | GO:0032552 | pyrimidine nucleotide binding(GO:0019103) deoxyribonucleotide binding(GO:0032552) |
| 0.4 | 3.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 1.2 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.4 | 79.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.4 | 3.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 0.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 2.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 4.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 4.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 1.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 7.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 3.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 1.9 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.3 | 1.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 2.4 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.3 | 8.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 10.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 3.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 4.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 1.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.3 | 2.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 5.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.3 | 2.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 7.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 4.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 2.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 3.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 0.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.0 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 2.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 5.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 3.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 3.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 2.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 5.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 2.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.2 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.9 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.2 | 5.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 4.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 7.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 1.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 0.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 12.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 2.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.1 | 2.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 3.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 3.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 7.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 7.7 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 1.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.5 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 1.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 1.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 13.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 12.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 8.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 2.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.6 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 3.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 4.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 2.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 1.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 9.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 3.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 2.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 3.1 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 1.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 17.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta-activated receptor activity(GO:0005024) transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 1.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.7 | 15.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.5 | 33.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 9.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 11.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 1.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 15.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 5.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 7.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 14.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 10.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 9.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 6.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 7.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 5.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 17.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 5.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 5.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 7.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 5.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 8.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 7.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 5.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 3.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 3.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 34.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 1.3 | 28.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 1.0 | 7.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 1.0 | 14.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.9 | 15.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.8 | 5.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.8 | 24.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.7 | 19.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.7 | 23.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 37.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.5 | 34.0 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.5 | 8.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.5 | 8.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 4.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.5 | 14.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.5 | 15.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 13.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.4 | 1.7 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.4 | 16.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 9.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.4 | 8.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.4 | 28.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 37.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 5.1 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.3 | 19.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 19.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 7.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 6.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 7.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.2 | 22.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 5.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 3.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 1.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 10.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 4.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 5.3 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.2 | 3.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 5.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 3.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 3.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 3.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 12.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 6.6 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 6.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 6.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 7.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 5.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.9 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |