Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
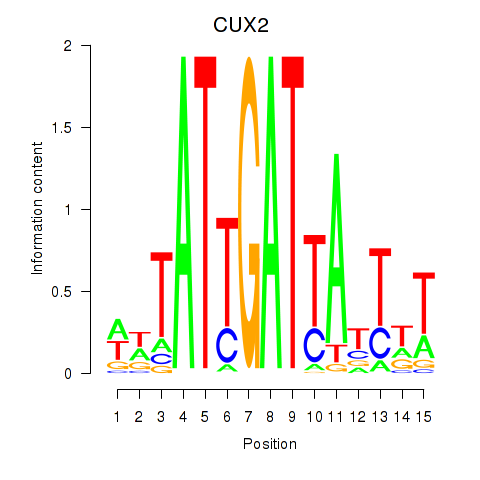
Results for CUX2
Z-value: 1.17
Transcription factors associated with CUX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX2
|
ENSG00000111249.14 | CUX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg38_v1_chr12_+_111034136_111034173 | 0.35 | 7.9e-08 | Click! |
Activity profile of CUX2 motif
Sorted Z-values of CUX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.7 | GO:1904884 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 4.8 | 23.8 | GO:1902952 | NMDA glutamate receptor clustering(GO:0097114) positive regulation of postsynaptic membrane organization(GO:1901628) positive regulation of dendritic spine maintenance(GO:1902952) |
| 4.3 | 13.0 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.3 | 12.9 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 4.0 | 11.9 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 4.0 | 11.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 3.9 | 35.4 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 3.9 | 11.6 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 3.7 | 11.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 3.4 | 44.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 3.2 | 13.0 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 3.2 | 12.7 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 2.7 | 8.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 2.7 | 8.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 2.5 | 7.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 2.4 | 7.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 2.2 | 6.5 | GO:0090155 | glucosylceramide biosynthetic process(GO:0006679) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 2.0 | 4.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.7 | 5.2 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 1.7 | 22.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.7 | 6.8 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.7 | 5.0 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 1.6 | 23.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.6 | 6.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.6 | 9.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.5 | 6.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.5 | 2.9 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.4 | 4.3 | GO:2001301 | cellular response to interleukin-13(GO:0035963) regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 1.4 | 4.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.4 | 9.8 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 1.4 | 4.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 1.4 | 4.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 1.4 | 8.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 1.3 | 19.8 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 1.3 | 5.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 1.3 | 6.3 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 1.2 | 11.7 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.2 | 11.5 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.1 | 3.4 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 1.0 | 3.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 1.0 | 4.9 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.9 | 9.5 | GO:0045176 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) apical protein localization(GO:0045176) |
| 0.9 | 8.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.9 | 7.9 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.8 | 11.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.8 | 4.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.8 | 13.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.8 | 7.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 3.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.7 | 6.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.7 | 2.2 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.7 | 11.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.7 | 2.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.7 | 14.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.7 | 4.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.7 | 4.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.7 | 2.8 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.7 | 8.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 5.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.7 | 12.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 2.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.6 | 3.2 | GO:0010807 | positive regulation of norepinephrine secretion(GO:0010701) regulation of synaptic vesicle priming(GO:0010807) |
| 0.6 | 6.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 1.9 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.6 | 4.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.6 | 2.9 | GO:0046439 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) homocysteine catabolic process(GO:0043418) L-cysteine metabolic process(GO:0046439) |
| 0.6 | 4.6 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.6 | 1.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.6 | 14.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.6 | 6.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.5 | 7.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.5 | 2.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.5 | 6.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.5 | 8.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.5 | 4.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 2.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 13.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.5 | 10.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.5 | 27.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 1.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 | 5.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.4 | 4.4 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 3.0 | GO:0046618 | drug export(GO:0046618) |
| 0.4 | 2.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 | 2.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 8.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 22.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.4 | 8.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 6.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.4 | 6.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.4 | 1.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.4 | 6.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 5.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.4 | 8.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 3.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 5.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 3.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.7 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 | 3.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 1.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.3 | 11.7 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.3 | 10.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.3 | 18.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 3.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 1.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 0.5 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.3 | 3.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.3 | 0.3 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.3 | 2.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 2.5 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 3.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 2.2 | GO:1901898 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 15.0 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.2 | 6.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 3.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 3.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 4.9 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 | 1.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 3.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 0.9 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.2 | 8.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 1.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 2.3 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.2 | 0.8 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.2 | 3.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.4 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 7.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 6.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 0.9 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.2 | 2.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 6.7 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 2.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 17.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 4.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 2.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 2.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 9.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 6.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 5.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 1.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.2 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.2 | 1.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 3.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 4.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 2.4 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.7 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 12.1 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 5.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 4.4 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 2.0 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.1 | 0.5 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.1 | 9.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 2.8 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 2.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 4.1 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 3.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.8 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 2.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 2.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 3.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 4.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 2.3 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 6.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 1.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 7.9 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 3.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 26.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 8.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 2.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 8.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 2.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 6.5 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.0 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 11.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 11.4 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.8 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 1.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 2.5 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 10.4 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) |
| 0.0 | 0.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 5.5 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 10.9 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 3.6 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 2.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 3.6 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 2.7 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 2.0 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 2.6 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 2.4 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0007292 | female gamete generation(GO:0007292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 3.4 | 44.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.6 | 13.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 2.2 | 6.7 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.2 | 3.7 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 1.2 | 8.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.0 | 38.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.0 | 4.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.9 | 11.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.8 | 3.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.8 | 7.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.8 | 5.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.8 | 15.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.7 | 17.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 10.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.6 | 5.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 9.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 5.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 2.5 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 3.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 3.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 14.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 4.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 77.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 1.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 21.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 14.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 15.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.3 | 6.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.3 | 0.8 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.3 | 3.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 4.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 12.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 23.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 6.9 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 8.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 4.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 3.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 19.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 2.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 4.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 1.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 4.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.2 | 20.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.2 | 6.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 3.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 6.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 9.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 3.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 8.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 15.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 9.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 7.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 6.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 14.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 14.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 11.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 6.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 81.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 3.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 7.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 7.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 4.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 7.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 5.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 110.9 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.4 | GO:0099572 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) postsynaptic specialization(GO:0099572) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 5.2 | 15.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 4.9 | 19.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 4.4 | 35.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.5 | 10.4 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 3.1 | 15.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 2.5 | 12.7 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 2.4 | 7.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 2.4 | 9.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.3 | 23.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 2.3 | 7.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 2.3 | 6.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 2.2 | 6.5 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 2.1 | 6.3 | GO:0035473 | lipase binding(GO:0035473) |
| 2.1 | 8.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.8 | 10.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 1.7 | 8.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.7 | 6.8 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 1.6 | 11.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 1.6 | 4.7 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.5 | 4.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.5 | 11.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.4 | 4.3 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 1.4 | 4.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.1 | 5.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 1.1 | 15.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.0 | 3.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.0 | 6.0 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.0 | 2.9 | GO:0098809 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.9 | 2.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.9 | 2.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.8 | 3.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.8 | 11.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.8 | 9.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 5.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.7 | 11.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.7 | 7.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.7 | 12.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.7 | 21.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 13.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 2.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.6 | 3.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.6 | 1.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.6 | 3.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.6 | 1.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.6 | 4.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 18.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 2.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 3.3 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.5 | 8.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 2.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.5 | 43.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 2.9 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.5 | 4.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.5 | 1.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.5 | 6.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 2.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.5 | 13.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.4 | 23.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.4 | 6.8 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 10.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 5.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 2.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 3.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 12.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 11.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 3.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 6.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 6.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 4.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 3.9 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 2.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 5.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 1.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 2.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 4.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 7.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 3.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 5.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 4.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 8.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 4.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 3.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 3.0 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.3 | 3.2 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.3 | 1.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 0.8 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.3 | 5.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 7.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 5.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 45.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.2 | 9.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 4.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 19.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 0.8 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.2 | 2.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 0.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 3.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 2.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 9.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 3.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 10.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 18.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 4.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 7.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 12.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.4 | GO:0097258 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 4.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 7.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 9.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 20.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 6.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 10.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 3.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 3.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 9.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 7.2 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 19.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 3.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 8.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 9.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 8.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 4.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 7.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.7 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 2.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 14.7 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 3.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 3.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.0 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 2.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.3 | GO:0001047 | core promoter binding(GO:0001047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 42.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 15.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 8.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 10.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 19.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 6.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 60.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 15.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 11.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 10.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 6.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 9.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 5.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 26.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 11.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 5.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 34.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 9.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 11.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 13.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 20.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 6.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 7.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 8.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 2.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 7.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 4.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 27.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 2.0 | 47.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 1.7 | 27.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 1.6 | 44.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.3 | 15.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.8 | 19.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.8 | 18.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 4.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 10.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 12.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 9.8 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.5 | 10.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 9.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 14.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 12.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 15.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 10.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 3.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 6.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 38.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 6.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 18.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 11.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 4.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 11.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 3.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 16.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 3.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 5.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 4.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 4.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 10.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 9.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 6.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 13.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 6.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 5.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |