Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for EP300
Z-value: 1.05
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.14 | EP300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg38_v1_chr22_+_41092585_41092633, hg38_v1_chr22_+_41092869_41093005 | -0.27 | 7.3e-05 | Click! |
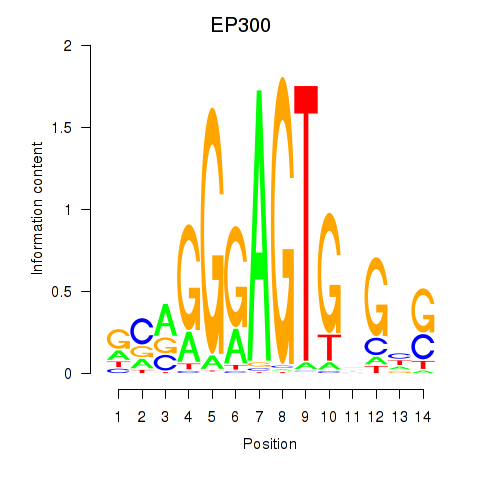
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 42.2 | GO:0030421 | defecation(GO:0030421) |
| 5.0 | 19.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 4.0 | 48.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 3.5 | 35.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 3.5 | 79.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 3.1 | 12.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 2.9 | 11.8 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 2.7 | 8.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 2.6 | 7.8 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 2.3 | 13.5 | GO:0060356 | leucine import(GO:0060356) |
| 2.1 | 6.2 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 2.0 | 6.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.9 | 19.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.7 | 5.2 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 1.7 | 5.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.7 | 14.9 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.5 | 6.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 1.5 | 11.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.4 | 12.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.3 | 2.7 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 1.3 | 3.8 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.2 | 4.8 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 1.2 | 10.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.1 | 7.9 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 1.0 | 3.1 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 1.0 | 7.8 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 1.0 | 2.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.8 | GO:1904434 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.9 | 6.5 | GO:0015866 | ADP transport(GO:0015866) |
| 0.9 | 3.7 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) prostate gland stromal morphogenesis(GO:0060741) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.9 | 2.8 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.9 | 3.4 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.8 | 2.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.8 | 6.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.8 | 12.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.8 | 5.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.8 | 5.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.8 | 7.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 2.9 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.7 | 5.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.7 | 2.8 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.6 | 4.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.6 | 9.4 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.6 | 10.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.6 | 11.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.6 | 9.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.6 | 1.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.6 | 1.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.6 | 6.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.5 | 4.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.5 | 2.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.5 | 2.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 1.9 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.5 | 5.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 6.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 11.5 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.4 | 1.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.4 | 1.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.4 | 11.7 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.4 | 3.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 2.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 2.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 | 4.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.4 | 1.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.4 | 2.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.4 | 1.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 7.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 4.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 3.0 | GO:0050713 | common-partner SMAD protein phosphorylation(GO:0007182) positive regulation of MHC class I biosynthetic process(GO:0045345) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.3 | 9.1 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.3 | 1.3 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.3 | 1.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.3 | 4.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 7.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.3 | 1.5 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.3 | 0.8 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 1.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.3 | 2.4 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.3 | 0.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 1.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 5.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 0.7 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 0.6 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.2 | 1.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 2.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 2.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.5 | GO:1902714 | antifungal humoral response(GO:0019732) negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.2 | 2.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 5.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 3.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 2.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.9 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.2 | 0.8 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.5 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.2 | 8.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 | 2.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 2.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 2.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.9 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 2.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.9 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 3.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.6 | GO:0061368 | axonogenesis involved in innervation(GO:0060385) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 3.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.6 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 9.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 1.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 2.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.4 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.1 | 1.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 6.4 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 6.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 2.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 3.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.9 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 6.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 4.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.7 | GO:0003360 | brainstem development(GO:0003360) ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 4.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.8 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 1.2 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 2.4 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 4.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.4 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 3.3 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 1.9 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 2.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.4 | GO:0097646 | positive regulation of protein kinase A signaling(GO:0010739) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.6 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.3 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 1.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 2.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.7 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 3.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.1 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 1.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 5.6 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 2.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 3.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 1.7 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0071907 | determination of pancreatic left/right asymmetry(GO:0035469) determination of digestive tract left/right asymmetry(GO:0071907) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 2.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.7 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.7 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.8 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 2.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0035404 | histone-serine phosphorylation(GO:0035404) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 35.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 3.1 | 79.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.3 | 48.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.2 | 19.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 1.0 | 9.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.0 | 6.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.8 | 5.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 7.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.8 | 4.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.7 | 7.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.7 | 6.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.7 | 5.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.7 | 6.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.7 | 7.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.6 | 7.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.6 | 1.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.6 | 19.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 2.9 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 10.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 12.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.5 | 2.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 1.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.4 | 3.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 1.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 5.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 5.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 12.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 2.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.3 | 4.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 7.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 31.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 1.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 3.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.2 | 4.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 11.0 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 4.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 22.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 1.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.7 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 1.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 3.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 0.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 2.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 2.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 5.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 6.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 15.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 5.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 5.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 4.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 13.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 7.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 3.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 5.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 9.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 9.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 2.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 18.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0044218 | growing cell tip(GO:0035838) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 5.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 1.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 35.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 3.4 | 10.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 2.9 | 11.7 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 2.8 | 79.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 2.7 | 19.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 2.4 | 12.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 2.1 | 12.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.5 | 12.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.5 | 11.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.2 | 21.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 1.0 | 13.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.0 | 5.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.9 | 6.5 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.8 | 11.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.8 | 4.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.8 | 2.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.8 | 7.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.8 | 7.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 5.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.7 | 2.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.7 | 7.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.6 | 1.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 3.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 6.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.6 | 2.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 1.7 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.5 | 4.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 5.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.5 | 4.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.5 | 1.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 1.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.4 | 1.9 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 1.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.4 | 1.9 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 2.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 3.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 1.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 6.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 2.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 1.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 2.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 8.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 6.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.3 | 7.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 2.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 5.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 6.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.3 | 6.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 4.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 6.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 43.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 40.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 1.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 6.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 0.9 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 5.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.7 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 5.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 10.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 6.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 16.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 3.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 3.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 7.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 20.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 2.5 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 2.9 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 2.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.0 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 2.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 18.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 20.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 1.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 3.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 2.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.4 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 1.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 4.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 1.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 4.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 3.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 5.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 7.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 121.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.6 | 49.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.4 | 57.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 7.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 1.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 13.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 5.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 3.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 5.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 28.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 14.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 6.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 10.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 3.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 7.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 9.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 8.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 16.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 89.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.6 | 21.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 35.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.5 | 7.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 17.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 19.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.4 | 53.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 12.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 7.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 6.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 6.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 4.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 7.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 16.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 4.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 3.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 10.4 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 3.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 6.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 8.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 3.5 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 2.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 5.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |