Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
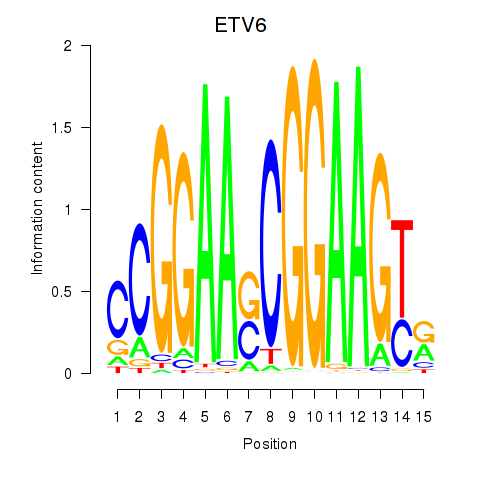
Results for ETV6
Z-value: 1.51
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.11 | ETV6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV6 | hg38_v1_chr12_+_11649666_11649751 | -0.24 | 3.9e-04 | Click! |
Activity profile of ETV6 motif
Sorted Z-values of ETV6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.5 | 58.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 12.7 | 76.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 9.8 | 29.3 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 9.4 | 37.5 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 9.1 | 36.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 8.6 | 25.7 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 8.4 | 42.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 7.9 | 31.7 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 6.8 | 6.8 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 6.2 | 86.6 | GO:0015074 | DNA integration(GO:0015074) |
| 5.1 | 25.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 5.1 | 40.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 5.0 | 30.2 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 4.9 | 14.7 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 4.7 | 135.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 4.4 | 22.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) pronephric nephron development(GO:0039019) |
| 4.4 | 39.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 3.9 | 11.7 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 3.8 | 11.5 | GO:1903774 | ubiquitin-dependent endocytosis(GO:0070086) positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 3.7 | 11.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 3.7 | 18.5 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 3.7 | 18.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 3.7 | 25.7 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 3.6 | 21.7 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 3.5 | 14.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 3.1 | 9.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 3.1 | 12.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 3.1 | 27.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 3.0 | 9.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 2.8 | 8.5 | GO:1901355 | response to rapamycin(GO:1901355) |
| 2.8 | 22.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 2.8 | 41.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 2.8 | 22.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 2.8 | 13.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 2.5 | 7.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 2.5 | 15.0 | GO:0019255 | UDP-glucuronate biosynthetic process(GO:0006065) glucose 1-phosphate metabolic process(GO:0019255) |
| 2.5 | 14.8 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 2.5 | 12.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 2.4 | 9.8 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 2.4 | 26.9 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 2.4 | 11.8 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 2.3 | 9.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 2.3 | 6.8 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 2.2 | 6.7 | GO:0034402 | mRNA export from nucleus in response to heat stress(GO:0031990) recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 2.2 | 13.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 2.2 | 6.6 | GO:1903719 | positive regulation of endodeoxyribonuclease activity(GO:0032079) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 2.2 | 6.5 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 2.1 | 16.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 2.0 | 10.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.8 | 10.8 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.7 | 22.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 1.7 | 5.0 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 1.6 | 9.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 1.6 | 17.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 1.6 | 15.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 1.6 | 28.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.5 | 27.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 1.5 | 12.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 1.5 | 12.1 | GO:1902661 | positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 1.5 | 22.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 1.5 | 4.5 | GO:0071336 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.5 | 4.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.5 | 5.8 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 1.5 | 13.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.4 | 13.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 1.4 | 51.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 1.4 | 4.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.4 | 26.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 1.4 | 12.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 1.3 | 10.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 1.3 | 29.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.2 | 23.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.2 | 12.8 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 1.2 | 13.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 1.1 | 10.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.1 | 14.7 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 1.1 | 17.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.1 | 4.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 1.1 | 12.9 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 1.0 | 7.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.0 | 16.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 1.0 | 14.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 1.0 | 17.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 1.0 | 6.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 1.0 | 7.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.0 | 8.8 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.0 | 5.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.9 | 36.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.9 | 10.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.9 | 7.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.9 | 7.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.8 | 8.4 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.8 | 2.5 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.8 | 11.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.8 | 9.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.8 | 11.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.8 | 2.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.8 | 8.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.8 | 17.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.8 | 6.8 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.8 | 96.6 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.8 | 24.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.7 | 8.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.7 | 6.5 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.7 | 88.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.7 | 58.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.7 | 10.3 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.7 | 45.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.7 | 3.4 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.6 | 19.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.6 | 12.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.6 | 7.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.6 | 9.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 8.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.6 | 1.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.6 | 18.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.6 | 21.2 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.6 | 4.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.6 | 4.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.6 | 13.0 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.5 | 6.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 17.9 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) |
| 0.5 | 7.6 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.5 | 8.0 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.5 | 6.8 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.5 | 5.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.5 | 6.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.5 | 8.7 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.5 | 9.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 5.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.5 | 0.9 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.5 | 9.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.4 | 7.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.4 | 8.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.4 | 12.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.4 | 7.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.4 | 1.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.4 | 8.8 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.4 | 4.0 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.4 | 4.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 28.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 32.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.4 | 1.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 6.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.4 | 1.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.4 | 13.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.3 | 1.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 1.0 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.3 | 2.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.3 | 2.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 5.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.3 | 7.7 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.3 | 1.0 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.3 | 5.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 2.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 4.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 3.6 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.3 | 4.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.3 | 3.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 13.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 5.9 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.2 | 13.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 8.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 3.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 17.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.2 | 3.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.2 | 0.9 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.2 | 10.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 5.8 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 2.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 3.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 6.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.2 | 2.0 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.2 | 0.6 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.2 | 1.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 0.9 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 8.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 10.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.2 | 10.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 1.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 9.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 7.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 6.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.1 | 4.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 7.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 43.8 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 1.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 5.1 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 24.8 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 2.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 4.2 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.3 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 2.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 3.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 2.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 2.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 6.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 2.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 7.0 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.7 | 141.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 9.4 | 37.5 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 8.5 | 25.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 7.5 | 30.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 7.4 | 29.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 7.1 | 70.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 6.0 | 42.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 5.8 | 29.0 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 5.8 | 40.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 4.6 | 13.8 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 4.5 | 13.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 4.0 | 36.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 3.7 | 11.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 3.3 | 66.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 3.2 | 31.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 3.1 | 15.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 3.0 | 14.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 2.9 | 14.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 2.9 | 95.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 2.8 | 8.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 2.6 | 10.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 2.6 | 13.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 2.5 | 70.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 2.4 | 31.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 2.4 | 7.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 2.3 | 6.8 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 2.2 | 28.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 2.2 | 26.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 2.1 | 10.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 2.1 | 25.7 | GO:0071203 | F-actin capping protein complex(GO:0008290) WASH complex(GO:0071203) |
| 2.1 | 38.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 1.9 | 9.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 1.9 | 26.9 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.9 | 17.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 1.9 | 22.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 1.7 | 5.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.5 | 12.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.5 | 5.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.4 | 17.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 1.3 | 6.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 1.3 | 6.5 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 1.3 | 6.5 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 1.3 | 11.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.3 | 12.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 1.3 | 8.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 1.2 | 6.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.2 | 26.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 1.2 | 4.7 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 1.2 | 46.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.1 | 13.8 | GO:0030897 | HOPS complex(GO:0030897) |
| 1.0 | 3.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.0 | 13.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 1.0 | 98.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.9 | 12.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.9 | 9.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.9 | 15.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.9 | 0.9 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.9 | 13.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.8 | 2.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.8 | 17.0 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.8 | 25.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.8 | 4.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.7 | 45.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.7 | 9.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.7 | 18.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.7 | 2.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.7 | 11.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 22.0 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.6 | 1.9 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.6 | 7.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.6 | 16.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.6 | 6.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.6 | 6.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.5 | 65.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.5 | 30.3 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 2.0 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.5 | 5.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 26.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.5 | 29.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 29.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.4 | 5.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.4 | 1.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 90.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.4 | 14.2 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.4 | 10.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.4 | 9.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 7.3 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.3 | 10.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 6.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 1.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 25.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 4.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 9.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.2 | 7.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.2 | 1.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 40.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.2 | 2.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.5 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 10.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.2 | 3.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 3.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 12.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 4.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 8.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 13.8 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 1.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 3.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 100.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 10.3 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.1 | 6.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 18.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.9 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 2.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 6.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 29.3 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 9.3 | 28.0 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 8.5 | 25.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 7.8 | 62.1 | GO:0070990 | snRNP binding(GO:0070990) |
| 7.0 | 42.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 6.1 | 36.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 5.9 | 64.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 5.4 | 21.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 5.3 | 26.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 4.4 | 39.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 4.4 | 13.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 4.1 | 29.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 4.0 | 15.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 3.9 | 11.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 3.7 | 15.0 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 3.7 | 37.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 3.7 | 14.7 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 3.4 | 10.2 | GO:0030622 | U4atac snRNA binding(GO:0030622) box C/D snoRNA binding(GO:0034512) |
| 3.2 | 9.6 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 3.1 | 9.4 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 3.0 | 9.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 3.0 | 14.8 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 2.8 | 8.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 2.8 | 13.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 2.6 | 10.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 2.6 | 13.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 2.6 | 28.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.5 | 17.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 2.5 | 10.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 2.5 | 7.4 | GO:0034416 | phosphohistidine phosphatase activity(GO:0008969) bisphosphoglycerate phosphatase activity(GO:0034416) inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 2.4 | 63.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 2.4 | 14.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 2.4 | 14.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 2.2 | 6.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 2.2 | 6.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 2.1 | 10.6 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 2.1 | 16.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 2.1 | 35.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 2.0 | 138.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 2.0 | 14.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.9 | 5.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.7 | 7.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 1.6 | 7.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.6 | 20.3 | GO:0046790 | virion binding(GO:0046790) |
| 1.5 | 4.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.4 | 7.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 1.4 | 12.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.4 | 14.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.3 | 6.5 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 1.3 | 25.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 1.2 | 12.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.1 | 14.7 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 1.1 | 7.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.1 | 5.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.0 | 7.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 1.0 | 6.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.0 | 2.9 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.0 | 26.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 14.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.9 | 13.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.9 | 5.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.9 | 26.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.9 | 13.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 6.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.8 | 45.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.8 | 4.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.7 | 4.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.7 | 8.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.7 | 21.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.7 | 6.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.7 | 21.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.7 | 18.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 12.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.7 | 141.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.6 | 11.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 8.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 9.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.6 | 18.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.6 | 3.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.6 | 5.7 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.6 | 8.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 6.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 24.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.5 | 1.6 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.5 | 22.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.5 | 97.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.5 | 31.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.5 | 10.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.5 | 2.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.5 | 10.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 4.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.5 | 2.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.5 | 20.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.5 | 15.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 10.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.5 | 22.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.5 | 0.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 27.6 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.4 | 22.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.4 | 7.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 99.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.4 | 11.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 9.1 | GO:0044548 | GTPase inhibitor activity(GO:0005095) S100 protein binding(GO:0044548) |
| 0.4 | 9.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 9.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 9.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 7.5 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.3 | 3.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 0.9 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.3 | 4.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 5.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 15.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 4.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 4.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 6.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 4.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 3.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.2 | 3.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 6.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 9.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 2.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.7 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 0.9 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.2 | 64.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.2 | 14.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 7.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 35.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 2.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 2.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 8.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 18.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 3.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 8.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 22.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 29.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 7.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 4.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 5.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 1.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 5.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 10.4 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 7.8 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 1.9 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.8 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 55.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.1 | 54.3 | PID MYC PATHWAY | C-MYC pathway |
| 1.1 | 65.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.0 | 26.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.7 | 11.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.6 | 29.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.6 | 17.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 23.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.5 | 11.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 5.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.4 | 14.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 25.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 8.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.3 | 35.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 9.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 15.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 15.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.3 | 10.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 13.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 11.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 14.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 1.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 7.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 7.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 9.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 8.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 6.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 6.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 95.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 7.5 | 141.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 4.5 | 67.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.6 | 39.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 2.4 | 33.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 2.2 | 92.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 2.2 | 166.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.9 | 44.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 1.7 | 58.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 1.6 | 25.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 1.6 | 14.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 1.5 | 13.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.5 | 179.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 1.3 | 21.2 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 1.2 | 17.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 1.1 | 40.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.0 | 12.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.9 | 67.8 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.8 | 19.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.8 | 11.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.7 | 21.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.7 | 49.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.7 | 21.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.7 | 11.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.6 | 15.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.6 | 15.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 14.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.5 | 48.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.5 | 20.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.5 | 13.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.5 | 10.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.5 | 17.2 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.5 | 11.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 14.8 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.5 | 12.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 32.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.4 | 6.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 15.0 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.4 | 27.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 32.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 9.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.4 | 8.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.4 | 6.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 11.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 10.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.3 | 4.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 8.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 7.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 5.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 1.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.2 | 5.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.5 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.2 | 4.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 9.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 3.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 3.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 11.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 3.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 4.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |