Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for EZH2
Z-value: 1.98
Transcription factors associated with EZH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EZH2
|
ENSG00000106462.12 | EZH2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EZH2 | hg38_v1_chr7_-_148883474_148883509 | -0.64 | 7.7e-27 | Click! |
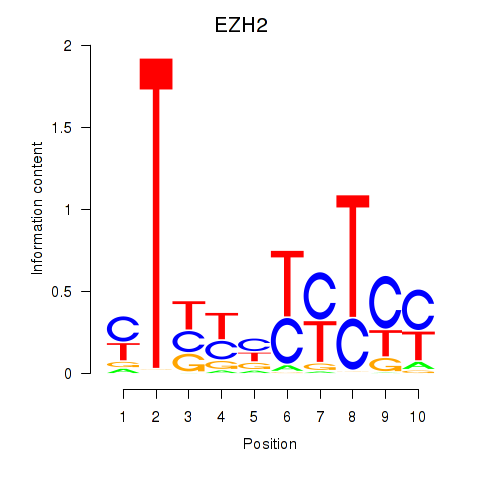
Activity profile of EZH2 motif
Sorted Z-values of EZH2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EZH2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 250.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 12.7 | 38.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 11.7 | 35.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 11.7 | 46.7 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 9.3 | 27.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 8.9 | 26.8 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 7.9 | 23.7 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 6.8 | 20.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 6.5 | 110.5 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 6.5 | 45.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 6.3 | 57.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 5.8 | 17.4 | GO:0042704 | uterine wall breakdown(GO:0042704) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 5.3 | 21.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 5.0 | 15.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 4.9 | 24.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 4.6 | 13.9 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 4.5 | 18.0 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 4.4 | 13.3 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 4.2 | 21.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 4.0 | 20.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 4.0 | 15.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 4.0 | 11.9 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 3.9 | 15.6 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 3.9 | 11.7 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 3.8 | 79.0 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 3.6 | 14.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 3.5 | 10.4 | GO:0060875 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 3.5 | 3.5 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 3.2 | 16.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 3.1 | 62.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 3.1 | 12.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 3.1 | 9.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 3.1 | 6.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 3.0 | 12.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 3.0 | 9.1 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 3.0 | 9.0 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 3.0 | 20.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 3.0 | 47.3 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 2.8 | 36.8 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 2.8 | 39.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 2.8 | 22.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 2.8 | 11.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 2.8 | 30.5 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 2.8 | 19.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 2.8 | 19.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 2.7 | 10.8 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 2.7 | 8.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.5 | 22.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 2.4 | 7.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 2.3 | 9.4 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 2.3 | 6.9 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 2.3 | 6.9 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 2.3 | 6.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 2.3 | 6.8 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 2.2 | 8.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 2.2 | 19.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 2.2 | 26.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.2 | 4.4 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 2.2 | 8.7 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 2.2 | 6.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 2.2 | 6.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 2.1 | 10.7 | GO:0010159 | specification of organ position(GO:0010159) |
| 2.1 | 16.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 2.0 | 30.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 2.0 | 6.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 1.9 | 5.8 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 1.9 | 28.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.8 | 11.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 1.8 | 49.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.8 | 5.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 1.8 | 20.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 1.8 | 7.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.8 | 12.5 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 1.8 | 12.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.8 | 12.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.8 | 8.9 | GO:0035645 | posterior midgut development(GO:0007497) enteric smooth muscle cell differentiation(GO:0035645) |
| 1.8 | 12.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 1.8 | 12.3 | GO:0045007 | depurination(GO:0045007) |
| 1.7 | 5.2 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 1.7 | 5.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.7 | 10.2 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 1.7 | 5.1 | GO:2001025 | cellular response to mycotoxin(GO:0036146) response to cyclosporin A(GO:1905237) positive regulation of response to drug(GO:2001025) |
| 1.7 | 6.7 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.7 | 15.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.7 | 3.3 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 1.7 | 14.9 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.6 | 6.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.6 | 48.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 1.5 | 3.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 1.5 | 20.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 1.5 | 4.4 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.4 | 18.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.4 | 10.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.4 | 8.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.4 | 7.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 1.4 | 4.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 1.4 | 7.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 1.4 | 8.4 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 1.4 | 61.6 | GO:0008038 | neuron recognition(GO:0008038) |
| 1.4 | 13.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.4 | 4.2 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.4 | 1.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 1.3 | 5.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.3 | 13.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.3 | 5.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.3 | 5.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 1.3 | 5.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.3 | 20.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 1.3 | 34.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 1.3 | 6.3 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 1.2 | 3.7 | GO:0071812 | regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 1.2 | 34.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 1.2 | 2.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.2 | 5.0 | GO:0030432 | peristalsis(GO:0030432) |
| 1.2 | 42.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.2 | 9.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.2 | 3.6 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 1.2 | 11.8 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 1.2 | 2.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 1.2 | 4.6 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 1.1 | 9.0 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.1 | 3.4 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 1.1 | 7.9 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.1 | 6.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.1 | 16.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 1.1 | 3.3 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 1.1 | 29.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.1 | 8.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.1 | 1.1 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 1.1 | 5.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 1.1 | 5.3 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 1.0 | 11.5 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 1.0 | 2.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 1.0 | 3.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.0 | 19.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.0 | 3.0 | GO:0033082 | extrathymic T cell differentiation(GO:0033078) regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 1.0 | 7.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 1.0 | 28.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 1.0 | 10.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.0 | 4.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 1.0 | 155.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 1.0 | 15.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.0 | 5.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 1.0 | 13.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.0 | 2.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 1.0 | 4.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.9 | 13.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.9 | 10.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 3.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.9 | 2.8 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.9 | 22.4 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.9 | 1.8 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.9 | 16.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.9 | 7.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.9 | 10.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.8 | 2.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.8 | 5.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.8 | 56.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.8 | 4.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.8 | 4.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.8 | 14.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.8 | 2.4 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.8 | 3.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.8 | 18.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.8 | 7.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.8 | 5.5 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.8 | 6.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.8 | 15.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 8.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.8 | 8.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.7 | 2.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.7 | 16.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.7 | 3.6 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 15.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.7 | 8.7 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.7 | 24.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.7 | 2.8 | GO:2000173 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.7 | 2.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.7 | 8.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.7 | 18.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.7 | 6.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.7 | 14.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.7 | 3.4 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.7 | 2.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.7 | 2.0 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.7 | 4.0 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.6 | 7.1 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 1.9 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 1.9 | GO:0045900 | negative regulation of translational elongation(GO:0045900) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.6 | 17.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.6 | 5.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.6 | 3.8 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.6 | 6.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.6 | 1.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.6 | 1.9 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.6 | 2.5 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.6 | 8.0 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.6 | 1.2 | GO:0002876 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.6 | 6.0 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.6 | 16.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.6 | 2.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.6 | 2.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 | 6.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.6 | 15.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 | 6.1 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.5 | 4.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.5 | 4.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.5 | 12.8 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.5 | 2.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.5 | 1.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.5 | 0.5 | GO:2000538 | positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.5 | 1.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 9.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.5 | 13.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.5 | 2.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.5 | 8.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.5 | 1.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.5 | 7.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 22.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.5 | 0.9 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) |
| 0.5 | 3.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 4.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.5 | 10.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.5 | 1.8 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 1.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.4 | 29.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.4 | 9.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.4 | 1.8 | GO:0002879 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 1.7 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.4 | 6.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.4 | 7.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.4 | 2.5 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.4 | 1.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 0.8 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.4 | 6.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.4 | 6.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 12.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.4 | 3.2 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.4 | 6.3 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.4 | 2.0 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.4 | 22.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.4 | 4.6 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.4 | 11.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.4 | 3.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.4 | 3.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 5.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.4 | 3.0 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 10.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.4 | 6.8 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.4 | 4.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 0.7 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.4 | 1.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.4 | 1.4 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.4 | 4.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 3.9 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.3 | 6.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 16.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 2.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 4.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.3 | 1.0 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.3 | 5.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.3 | 2.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 11.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.3 | 5.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 2.8 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 3.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 14.2 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.3 | 3.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 6.1 | GO:0030826 | regulation of cGMP biosynthetic process(GO:0030826) |
| 0.3 | 0.9 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.3 | 2.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 5.0 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.3 | 9.4 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.3 | 0.6 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.3 | 18.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.3 | 1.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.3 | 5.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.3 | 0.8 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.3 | 12.6 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.3 | 3.6 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.3 | 3.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.3 | 7.7 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.3 | 1.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 3.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 2.1 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.3 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 0.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.3 | 3.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.3 | 3.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 16.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.3 | 2.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.3 | 3.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 0.5 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.3 | 2.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 6.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.7 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.2 | 12.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.2 | 1.6 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 8.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.2 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 2.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 4.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 29.5 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.2 | 6.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 1.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 6.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 1.3 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.2 | 0.9 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 0.8 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.2 | 1.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 6.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 0.5 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 2.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 12.5 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.2 | 0.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 2.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 0.3 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 1.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 8.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 16.5 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.2 | 3.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 16.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 1.0 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.2 | 3.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.2 | 2.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 2.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 6.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 4.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 2.7 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.2 | 11.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.6 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.1 | 7.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 6.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 6.4 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 0.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 1.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 2.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 5.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 3.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 2.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.4 | GO:1902462 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 1.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.6 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.1 | 2.6 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 1.7 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.3 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 2.6 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 3.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 2.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.4 | GO:0098911 | regulation of ventricular cardiac muscle cell action potential(GO:0098911) |
| 0.1 | 5.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 3.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 12.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.9 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 1.0 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 0.7 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 4.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 2.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.3 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 3.8 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 27.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.6 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.1 | 5.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 1.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 279.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 12.0 | 36.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 11.8 | 58.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 9.5 | 57.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 7.9 | 23.7 | GO:0060987 | lipid tube(GO:0060987) |
| 6.2 | 18.7 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 5.5 | 99.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 4.8 | 19.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 4.4 | 17.7 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 4.0 | 11.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 2.7 | 21.8 | GO:0045179 | apical cortex(GO:0045179) |
| 2.7 | 16.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 2.6 | 65.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 2.6 | 36.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 2.5 | 32.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.1 | 6.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.8 | 14.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.8 | 10.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 1.7 | 3.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.7 | 8.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.6 | 21.0 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 1.5 | 7.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.5 | 6.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 1.4 | 7.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.4 | 34.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.4 | 4.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.3 | 16.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.3 | 25.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.3 | 20.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.2 | 6.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.1 | 4.6 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 1.1 | 6.8 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.1 | 11.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 11.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.1 | 3.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.0 | 9.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.0 | 8.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.0 | 235.0 | GO:0030426 | growth cone(GO:0030426) |
| 1.0 | 8.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.0 | 6.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 1.0 | 6.9 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 1.0 | 5.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.0 | 6.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.9 | 5.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 11.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.9 | 9.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.9 | 4.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 12.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.8 | 3.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.8 | 12.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.8 | 6.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 5.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.7 | 7.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 120.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.7 | 28.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.7 | 3.4 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 2.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 7.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 9.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.5 | 5.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.5 | 2.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.5 | 32.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.5 | 22.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.5 | 5.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 10.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 4.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.4 | 18.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.4 | 2.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 3.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 37.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 35.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 4.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.4 | 4.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 9.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 7.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 6.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 15.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 1.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.4 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 4.7 | GO:0031618 | chromocenter(GO:0010369) nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 25.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.4 | 4.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 6.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 4.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 4.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 6.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 3.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 22.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 1.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 19.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.3 | 1.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 0.6 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.3 | 37.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 11.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 7.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 3.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 6.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 8.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 4.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 15.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 2.7 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.2 | 2.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 4.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 25.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 2.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 3.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 47.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 2.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 3.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 64.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 5.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 5.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 4.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 8.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 8.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 5.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.2 | 3.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 7.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 5.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 5.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0070160 | occluding junction(GO:0070160) |
| 0.1 | 20.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 1.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 10.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 6.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 5.1 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 1.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 8.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 10.3 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 5.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 4.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.8 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 11.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 16.1 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 3.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.6 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 5.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 33.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 4.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 2.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 44.1 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 257.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 7.9 | 23.8 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 6.3 | 56.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 6.3 | 81.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 6.1 | 30.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 6.0 | 24.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 5.8 | 23.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 5.4 | 48.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 5.3 | 42.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 4.8 | 14.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 4.8 | 19.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 4.7 | 18.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 4.6 | 13.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 4.6 | 27.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 4.2 | 12.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 4.1 | 32.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 4.0 | 11.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 4.0 | 11.9 | GO:0030305 | heparanase activity(GO:0030305) |
| 4.0 | 47.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.9 | 11.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 3.9 | 31.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.5 | 17.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 3.4 | 13.7 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 3.4 | 27.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 3.3 | 20.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 3.3 | 65.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 3.2 | 15.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 3.1 | 18.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 3.1 | 12.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 3.1 | 12.3 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 2.9 | 17.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 2.8 | 8.4 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 2.8 | 19.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 2.7 | 8.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 2.6 | 18.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 2.6 | 20.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.6 | 7.7 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 2.6 | 15.4 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 2.5 | 20.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 2.5 | 10.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 2.2 | 13.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.1 | 19.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 2.1 | 8.3 | GO:0070728 | leucine binding(GO:0070728) |
| 2.1 | 64.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 2.0 | 18.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 2.0 | 28.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.0 | 18.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 2.0 | 25.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 2.0 | 5.9 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 1.9 | 5.8 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 1.9 | 5.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.9 | 21.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.8 | 9.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.8 | 7.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.8 | 14.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 1.7 | 17.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.7 | 5.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.7 | 35.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.7 | 6.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.6 | 4.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.6 | 22.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 1.6 | 11.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.5 | 10.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.5 | 10.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.4 | 39.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 1.4 | 10.0 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.4 | 7.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.4 | 2.8 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.4 | 5.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 1.4 | 4.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.3 | 25.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.3 | 9.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.3 | 15.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 1.3 | 5.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.3 | 45.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.2 | 14.6 | GO:0008430 | selenium binding(GO:0008430) |
| 1.2 | 15.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 1.2 | 21.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 1.2 | 10.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.2 | 24.2 | GO:0005112 | Notch binding(GO:0005112) |
| 1.1 | 12.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 1.1 | 10.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.1 | 11.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.1 | 24.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.1 | 3.3 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 1.1 | 10.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.1 | 8.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.0 | 9.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.0 | 5.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.0 | 11.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.0 | 15.1 | GO:0031432 | titin binding(GO:0031432) |
| 1.0 | 11.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 1.0 | 17.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.0 | 6.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.9 | 2.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.9 | 3.6 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.9 | 6.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.9 | 18.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.9 | 9.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.9 | 4.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.9 | 15.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.8 | 6.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.8 | 9.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.8 | 3.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 7.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.8 | 2.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.8 | 6.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.8 | 6.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 3.7 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 6.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.7 | 4.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.7 | 5.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.7 | 5.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.7 | 6.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.7 | 6.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.7 | 2.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 7.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.7 | 9.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.6 | 4.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 8.4 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.6 | 5.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 3.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 1.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 35.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 14.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 16.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.6 | 13.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.6 | 4.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 5.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.6 | 1.8 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.6 | 2.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.6 | 12.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 1.7 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.6 | 3.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.6 | 10.6 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.6 | 43.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 8.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 2.1 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) guanyl deoxyribonucleotide binding(GO:0032560) dGTP binding(GO:0032567) |
| 0.5 | 0.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 3.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 3.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.5 | 2.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.5 | 2.4 | GO:0031711 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.5 | 0.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.5 | 5.7 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.5 | 11.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 4.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 6.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 1.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.4 | 2.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 39.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 4.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 26.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.4 | 5.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 7.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 48.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.4 | 4.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 10.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 2.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.4 | 2.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 17.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.4 | 4.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 1.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 30.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 1.4 | GO:0032934 | sterol binding(GO:0032934) |
| 0.4 | 4.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 1.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.3 | 4.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.3 | 5.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 18.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 1.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 5.1 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.3 | 27.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 4.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 4.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 5.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 18.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 16.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 2.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 3.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.3 | 24.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 5.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 7.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.3 | 2.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 13.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 4.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 4.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 30.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 202.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.3 | 12.8 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.2 | 2.0 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.7 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.2 | 6.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.8 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 25.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 6.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 5.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 19.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.2 | 2.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 6.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 3.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 5.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 7.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 7.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 8.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 2.6 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 6.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 3.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 2.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 11.1 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 5.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 0.7 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 46.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 2.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 4.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 5.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 6.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 23.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 7.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.3 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 1.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 3.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.7 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 5.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 3.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 2.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 5.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 14.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 11.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 21.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 3.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.0 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 108.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.1 | 6.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 1.0 | 50.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.9 | 40.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.8 | 6.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.7 | 31.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.6 | 16.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.6 | 10.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.6 | 40.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.5 | 8.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.4 | 29.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.4 | 28.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 5.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 5.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 4.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 47.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 20.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 5.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 10.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 5.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 62.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 15.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 7.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 15.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 3.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 8.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 16.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 16.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 30.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 19.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 9.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 3.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 6.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 2.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 14.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 4.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 6.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 6.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 11.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 8.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 2.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 4.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 7.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 10.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 2.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 3.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 11.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 6.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 12.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 25.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 12.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 11.4 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 118.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 2.1 | 42.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.5 | 38.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.3 | 6.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 1.3 | 27.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 1.1 | 7.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.1 | 21.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 1.0 | 42.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.9 | 25.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.9 | 34.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 31.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.9 | 35.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.9 | 52.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.8 | 18.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.7 | 5.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 7.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.6 | 15.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.6 | 14.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.6 | 12.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.6 | 10.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 18.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.6 | 2.8 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.6 | 24.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 13.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 9.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.5 | 11.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 7.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.5 | 18.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 6.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 8.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.5 | 1.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.5 | 9.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.5 | 4.0 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.5 | 9.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 15.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.4 | 4.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 30.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 19.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 2.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.4 | 13.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 5.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 8.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 22.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 67.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 5.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 47.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 6.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.3 | 4.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 12.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 7.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 11.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 3.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 5.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 3.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.3 | 4.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 10.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 10.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 23.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 4.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 8.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 13.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 21.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 1.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 6.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 2.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 1.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 6.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.2 | 11.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 4.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 7.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 4.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 13.1 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 0.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 15.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 4.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 2.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |