Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
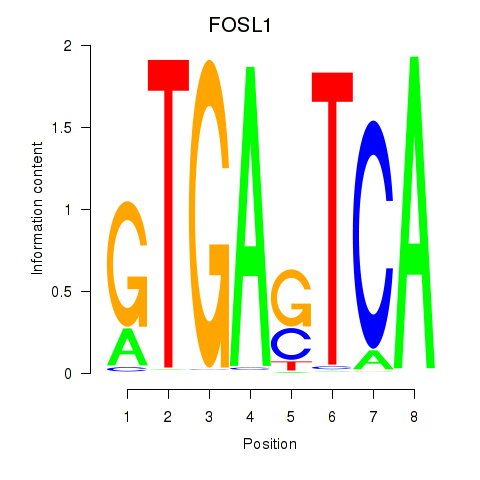
Results for FOSL1
Z-value: 1.47
Transcription factors associated with FOSL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL1
|
ENSG00000175592.9 | FOSL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL1 | hg38_v1_chr11_-_65900413_65900424 | 0.66 | 7.7e-29 | Click! |
Activity profile of FOSL1 motif
Sorted Z-values of FOSL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.5 | 129.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 14.7 | 73.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 13.4 | 40.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 11.4 | 45.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 9.4 | 56.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 8.3 | 24.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 8.2 | 41.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 7.4 | 96.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 6.8 | 20.3 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 5.9 | 23.6 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 5.3 | 90.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 5.2 | 20.9 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 5.1 | 30.3 | GO:0048669 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 4.3 | 21.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 4.2 | 42.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 4.0 | 56.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 3.9 | 11.8 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 3.9 | 19.6 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 3.8 | 11.3 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 3.4 | 37.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 3.3 | 9.8 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 3.2 | 12.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 3.0 | 9.0 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 2.8 | 16.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.7 | 10.8 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 2.5 | 12.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 2.5 | 12.3 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 2.4 | 34.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 2.4 | 7.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 2.3 | 4.6 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 2.3 | 29.5 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 2.3 | 9.0 | GO:1990637 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 2.2 | 9.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 2.2 | 42.2 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 2.2 | 6.6 | GO:0042946 | glucoside transport(GO:0042946) |
| 2.1 | 6.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.1 | 12.7 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 2.1 | 16.7 | GO:0010138 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 2.1 | 6.2 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 2.0 | 6.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.9 | 42.8 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 1.9 | 37.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 1.8 | 5.5 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 1.8 | 7.2 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 1.8 | 27.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.7 | 48.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.6 | 4.8 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 1.6 | 17.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.5 | 27.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.5 | 12.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.4 | 15.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 1.4 | 137.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 1.3 | 9.3 | GO:0030421 | defecation(GO:0030421) |
| 1.3 | 6.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 1.3 | 4.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.3 | 11.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 1.3 | 7.8 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 1.3 | 14.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.3 | 6.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.2 | 10.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.2 | 8.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.2 | 15.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.1 | 2.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.1 | 4.3 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 1.1 | 5.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.1 | 19.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.0 | 4.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.0 | 7.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 1.0 | 3.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.0 | 12.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 11.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.9 | 23.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.8 | 10.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.8 | 15.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.8 | 16.5 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.8 | 3.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.8 | 11.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.8 | 5.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.8 | 5.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.7 | 1.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.7 | 25.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.7 | 7.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.7 | 6.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 | 9.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.7 | 3.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.7 | 36.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.7 | 5.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.6 | 5.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 3.8 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.6 | 6.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.6 | 19.4 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.6 | 14.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.6 | 4.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.6 | 6.3 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.6 | 20.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.5 | 3.6 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.5 | 2.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.4 | 1.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.4 | 2.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 3.9 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 5.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.4 | 58.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.4 | 7.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.4 | 3.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 26.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.4 | 1.4 | GO:0052330 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.3 | 5.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 14.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 7.8 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.3 | 1.7 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.3 | 6.9 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.3 | 2.9 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.3 | 27.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 0.9 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.3 | 2.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 7.7 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.3 | 14.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 9.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 18.3 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.3 | 3.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.3 | 14.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 17.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 6.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.7 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 12.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 5.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 6.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 1.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 34.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 5.0 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.2 | 6.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 1.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 8.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 5.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 5.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 5.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 2.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 11.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 7.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.2 | 1.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) DNA cytosine deamination(GO:0070383) negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.2 | 4.5 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 10.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 2.4 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 7.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 6.7 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 4.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 3.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:0031342 | negative regulation of cell killing(GO:0031342) |
| 0.1 | 4.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.5 | GO:2000286 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 3.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 9.2 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.1 | 2.6 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.1 | 0.6 | GO:0014010 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell proliferation(GO:0014010) |
| 0.1 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 4.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 2.5 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.4 | GO:0002251 | organ or tissue specific immune response(GO:0002251) |
| 0.1 | 4.5 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 2.4 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 5.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 2.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 3.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 129.2 | GO:0005638 | lamin filament(GO:0005638) |
| 8.8 | 96.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 7.6 | 22.7 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 7.2 | 21.6 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 6.8 | 20.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 5.9 | 23.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 5.6 | 56.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 5.5 | 16.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 5.3 | 31.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 4.9 | 79.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 4.2 | 29.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 3.4 | 30.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 3.2 | 6.4 | GO:0043256 | laminin complex(GO:0043256) |
| 3.1 | 39.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 3.0 | 33.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 3.0 | 9.0 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 2.6 | 56.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 2.3 | 20.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 2.1 | 25.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.1 | 6.3 | GO:0044393 | microspike(GO:0044393) |
| 1.9 | 36.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 1.9 | 20.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.8 | 9.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.8 | 12.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.7 | 20.8 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.7 | 5.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.6 | 9.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.6 | 4.8 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 1.6 | 10.9 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.5 | 49.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 1.4 | 62.3 | GO:0032420 | stereocilium(GO:0032420) |
| 1.3 | 6.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.2 | 35.3 | GO:0030057 | desmosome(GO:0030057) |
| 1.2 | 10.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.2 | 23.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.1 | 15.8 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.1 | 35.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 1.1 | 127.7 | GO:0005604 | basement membrane(GO:0005604) |
| 1.0 | 9.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.0 | 27.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.9 | 16.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.8 | 13.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.8 | 2.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.7 | 10.9 | GO:0097433 | dense body(GO:0097433) |
| 0.7 | 6.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.7 | 16.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.6 | 7.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 10.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 58.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.5 | 4.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.5 | 7.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 3.8 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.4 | 6.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.4 | 15.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 38.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 3.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 5.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 5.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 12.8 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 32.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 18.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 9.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 20.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 12.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 0.9 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 9.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 3.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 9.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 5.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 13.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 20.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 4.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 12.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 7.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 6.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 8.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 16.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 4.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 25.1 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 15.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 5.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 6.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 7.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 8.9 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 36.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 6.5 | 19.6 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 5.9 | 29.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 5.9 | 23.6 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 3.4 | 23.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 3.3 | 9.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 3.1 | 31.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 3.1 | 21.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 3.0 | 30.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 3.0 | 9.0 | GO:0002135 | CTP binding(GO:0002135) |
| 2.8 | 45.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 2.8 | 16.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 2.7 | 107.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 2.6 | 99.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 2.5 | 12.7 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 2.5 | 9.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 2.4 | 14.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 2.2 | 6.6 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 2.2 | 10.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 2.1 | 14.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.0 | 20.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 2.0 | 16.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.9 | 44.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 1.9 | 20.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.8 | 5.5 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 1.7 | 11.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 1.5 | 12.0 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.5 | 41.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.4 | 7.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 1.4 | 12.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.3 | 4.0 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.3 | 73.4 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.2 | 18.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.2 | 7.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.2 | 23.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.2 | 22.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.1 | 14.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.1 | 34.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.1 | 5.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.0 | 11.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.0 | 3.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.0 | 7.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.0 | 37.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 1.0 | 27.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.9 | 80.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.9 | 9.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.9 | 3.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.9 | 6.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.9 | 9.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 5.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.9 | 6.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.8 | 12.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.8 | 10.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.8 | 4.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.7 | 10.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.7 | 18.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.7 | 8.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.7 | 35.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.7 | 11.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.6 | 5.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 14.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.6 | 14.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.6 | 6.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 2.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.6 | 14.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.6 | 8.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 5.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 5.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.6 | 3.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.5 | 4.6 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.5 | 10.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.5 | 23.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 2.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 1.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.5 | 3.8 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.5 | 1.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.5 | 79.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.5 | 7.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.4 | 18.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 6.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 22.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 7.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 18.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 22.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.3 | 4.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 35.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 0.9 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.3 | 3.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 9.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 13.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 16.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 6.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 17.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 31.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 3.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 120.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.2 | 3.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 6.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 59.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 7.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 5.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 5.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 28.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 3.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 6.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 2.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 2.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 64.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 2.4 | 90.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 2.0 | 23.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 1.7 | 76.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 1.7 | 37.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.7 | 44.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 1.4 | 50.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 1.4 | 66.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.3 | 139.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 1.2 | 95.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 1.2 | 62.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.0 | 76.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.8 | 49.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.6 | 26.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.5 | 39.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.5 | 34.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 10.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.5 | 21.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.4 | 21.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 9.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 71.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 7.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 19.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 7.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 2.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 6.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 4.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 5.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 23.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 7.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 12.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 23.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 16.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 96.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 2.7 | 56.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 2.5 | 129.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 2.2 | 105.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.9 | 137.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.8 | 16.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 1.5 | 63.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.4 | 49.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.4 | 35.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.4 | 27.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 1.2 | 40.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.1 | 16.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.1 | 5.5 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 1.1 | 45.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.0 | 45.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 1.0 | 23.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.9 | 25.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.8 | 9.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.7 | 9.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 4.6 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.6 | 18.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.6 | 10.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.6 | 4.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.6 | 11.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.6 | 50.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.5 | 8.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 6.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 12.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 5.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 42.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 11.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 16.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 6.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 2.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 10.0 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.2 | 3.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 5.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 4.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 6.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 5.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 11.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.6 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 11.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 9.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 9.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 11.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 4.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 5.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 8.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |