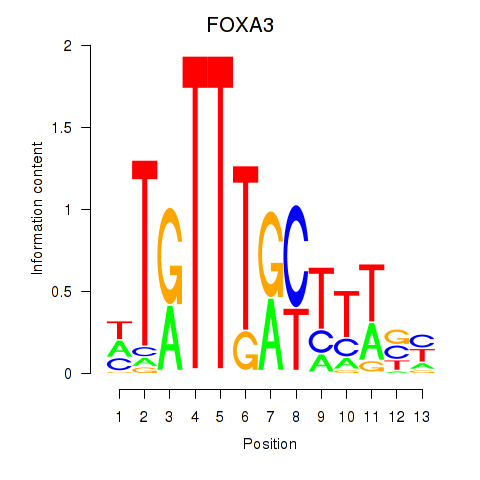
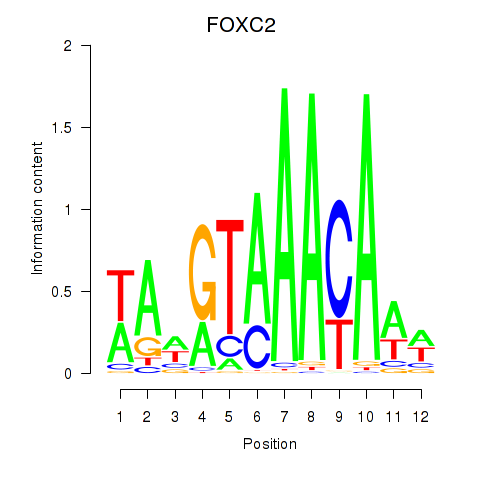
chr17_-_41528293
Show fit
13.87
ENST00000455635.1
ENST00000361566.7
KRT19
keratin 19
chr5_+_136058849
Show fit
12.04
ENST00000508076.5
TGFBI
transforming growth factor beta induced
chr18_+_3449620
Show fit
10.82
ENST00000405385.7
TGIF1
TGFB induced factor homeobox 1
chr7_+_134891566
Show fit
9.53
ENST00000424922.5
ENST00000495522.1
CALD1
caldesmon 1
chr8_+_96584920
Show fit
9.20
ENST00000521590.5
SDC2
syndecan 2
chr11_-_116837586
Show fit
8.92
ENST00000375320.5
ENST00000359492.6
ENST00000375329.6
ENST00000375323.5
ENST00000236850.5
APOA1
apolipoprotein A1
chr17_-_28368012
Show fit
8.65
ENST00000555059.2
ENSG00000273171.1
novel protein, readthrough between VTN and SEBOX
chr7_+_112423137
Show fit
8.56
ENST00000005558.8
ENST00000621379.4
IFRD1
interferon related developmental regulator 1
chr11_+_62419025
Show fit
8.40
ENST00000278282.3
SCGB1A1
secretoglobin family 1A member 1
chr12_+_12891554
Show fit
8.33
ENST00000014914.6
GPRC5A
G protein-coupled receptor class C group 5 member A
chr2_-_105438503
Show fit
8.29
ENST00000393352.7
ENST00000607522.1
FHL2
four and a half LIM domains 2
chr7_+_134779625
Show fit
8.28
ENST00000454108.5
ENST00000361675.7
CALD1
caldesmon 1
chr7_+_134779663
Show fit
8.25
ENST00000361901.6
CALD1
caldesmon 1
chr11_+_126327863
Show fit
7.58
ENST00000648516.1
DCPS
decapping enzyme, scavenger
chr10_-_116273009
Show fit
7.45
ENST00000439649.8
ENST00000369234.5
ENST00000682194.1
ENST00000355422.11
GFRA1
GDNF family receptor alpha 1
chr3_+_141387801
Show fit
7.11
ENST00000514251.5
ZBTB38
zinc finger and BTB domain containing 38
chr15_+_96325935
Show fit
7.02
ENST00000421109.6
NR2F2
nuclear receptor subfamily 2 group F member 2
chr11_-_108593738
Show fit
6.99
ENST00000525344.5
ENST00000265843.9
EXPH5
exophilin 5
chr7_-_108003122
Show fit
6.90
ENST00000393559.2
ENST00000222399.11
ENST00000676777.1
ENST00000439976.6
ENST00000393560.5
ENST00000677793.1
ENST00000679244.1
LAMB1
laminin subunit beta 1
chr2_+_108607140
Show fit
6.72
ENST00000410093.5
LIMS1
LIM zinc finger domain containing 1
chr10_-_125160499
Show fit
6.68
ENST00000494626.6
ENST00000337195.9
CTBP2
C-terminal binding protein 2
chr15_+_80072559
Show fit
6.34
ENST00000560228.5
ENST00000559835.5
ENST00000559775.5
ENST00000558688.5
ENST00000560392.5
ENST00000560976.5
ENST00000558272.5
ENST00000558390.5
ZFAND6
Links
zinc finger AN1-type containing 6
chr10_+_69801892
Show fit
6.18
ENST00000398978.8
ENST00000645393.2
ENST00000354547.7
ENST00000674121.1
ENST00000673842.1
ENST00000520267.5
COL13A1
collagen type XIII alpha 1 chain
chr21_-_26843012
Show fit
6.05
ENST00000517777.6
ADAMTS1
ADAM metallopeptidase with thrombospondin type 1 motif 1
chrX_-_41589970
Show fit
5.98
ENST00000378179.9
CASK
calcium/calmodulin dependent serine protein kinase
chr3_+_172040554
Show fit
5.95
ENST00000336824.8
ENST00000423424.5
FNDC3B
fibronectin type III domain containing 3B
chr21_-_26843063
Show fit
5.64
ENST00000678221.1
ADAMTS1
ADAM metallopeptidase with thrombospondin type 1 motif 1
chr2_+_73892967
Show fit
5.62
ENST00000409731.7
ENST00000409918.5
ENST00000442912.5
ENST00000345517.8
ENST00000409624.1
ACTG2
actin gamma 2, smooth muscle
chr8_-_63026179
Show fit
5.57
ENST00000677919.1
GGH
gamma-glutamyl hydrolase
chr7_+_48924559
Show fit
5.56
ENST00000650262.1
CDC14C
cell division cycle 14C
chr18_-_28036585
Show fit
5.44
ENST00000399380.7
CDH2
cadherin 2
chr10_-_125161019
Show fit
5.43
ENST00000411419.6
CTBP2
C-terminal binding protein 2
chr3_+_142596385
Show fit
5.29
ENST00000457734.7
ENST00000483373.5
ENST00000475296.5
ENST00000495744.5
ENST00000476044.5
ENST00000461644.5
ENST00000464320.5
PLS1
plastin 1
chr5_+_96663010
Show fit
5.29
ENST00000506811.5
ENST00000514055.5
ENST00000508608.6
CAST
calpastatin
chr1_-_56966133
Show fit
5.23
ENST00000535057.5
ENST00000543257.5
C8B
complement C8 beta chain
chr17_-_48610971
Show fit
5.17
ENST00000239165.9
HOXB7
homeobox B7
chr6_+_52420992
Show fit
5.12
ENST00000636954.1
ENST00000636566.1
ENST00000638075.1
EFHC1
EF-hand domain containing 1
chr10_+_122163672
Show fit
5.11
ENST00000369004.7
ENST00000260733.7
TACC2
transforming acidic coiled-coil containing protein 2
chr3_+_69936583
Show fit
5.01
ENST00000314557.10
ENST00000394351.9
MITF
melanocyte inducing transcription factor
chr10_+_122163590
Show fit
5.00
ENST00000368999.5
TACC2
transforming acidic coiled-coil containing protein 2
chr5_-_138338325
Show fit
4.99
ENST00000510119.1
ENST00000513970.5
CDC25C
cell division cycle 25C
chr9_+_68241854
Show fit
4.91
ENST00000616550.4
ENST00000618217.4
ENST00000377342.9
ENST00000478048.5
ENST00000360171.11
CBWD3
COBW domain containing 3
chr14_-_24195334
Show fit
4.89
ENST00000530563.1
ENST00000528895.5
ENST00000528669.5
ENST00000532632.1
ENST00000261789.9
TM9SF1
transmembrane 9 superfamily member 1
chr1_-_150697128
Show fit
4.89
ENST00000427665.1
ENST00000271732.8
GOLPH3L
golgi phosphoprotein 3 like
chr9_-_41189310
Show fit
4.76
ENST00000456520.5
ENST00000377391.8
ENST00000613716.4
ENST00000617933.1
CBWD6
COBW domain containing 6
chr15_-_58749569
Show fit
4.71
ENST00000402627.5
ENST00000559053.1
ENST00000260408.8
ENST00000561288.1
ENST00000461408.2
ENST00000439637.5
ENST00000558004.1
ADAM10
ADAM metallopeptidase domain 10
chr5_+_32531786
Show fit
4.71
ENST00000512913.5
SUB1
SUB1 regulator of transcription
chr4_+_146175702
Show fit
4.59
ENST00000296581.11
ENST00000649747.1
ENST00000502781.5
LSM6
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated
chr4_-_139302460
Show fit
4.57
ENST00000394223.2
ENST00000676245.1
NDUFC1
NADH:ubiquinone oxidoreductase subunit C1
chr4_-_139302516
Show fit
4.55
ENST00000394228.5
ENST00000539387.5
NDUFC1
NADH:ubiquinone oxidoreductase subunit C1
chr6_+_36676489
Show fit
4.49
ENST00000448526.6
CDKN1A
cyclin dependent kinase inhibitor 1A
chr5_+_96662969
Show fit
4.36
ENST00000514845.5
ENST00000675663.1
CAST
calpastatin
chrX_-_20218941
Show fit
4.31
ENST00000457145.6
RPS6KA3
ribosomal protein S6 kinase A3
chr18_+_49562049
Show fit
4.28
ENST00000261292.9
ENST00000427224.6
ENST00000580036.5
LIPG
lipase G, endothelial type
chr11_-_11353241
Show fit
4.12
ENST00000528848.3
CSNK2A3
casein kinase 2 alpha 3
chr2_+_113437691
Show fit
4.06
ENST00000259199.9
ENST00000416503.6
ENST00000433343.6
CBWD2
COBW domain containing 2
chrX_-_41665766
Show fit
4.03
ENST00000643043.2
ENST00000486402.1
ENST00000646087.2
CASK
calcium/calmodulin dependent serine protein kinase
chr17_-_19387170
Show fit
4.00
ENST00000395592.6
ENST00000299610.5
MFAP4
microfibril associated protein 4
chr3_-_108222383
Show fit
3.98
ENST00000264538.4
IFT57
intraflagellar transport 57
chr14_+_55661272
Show fit
3.97
ENST00000555573.5
KTN1
kinectin 1
chr3_-_134250831
Show fit
3.91
ENST00000623711.4
RYK
receptor like tyrosine kinase
chr11_+_102112445
Show fit
3.87
ENST00000524575.5
YAP1
Yes1 associated transcriptional regulator
chr7_+_134891400
Show fit
3.79
ENST00000393118.6
CALD1
caldesmon 1
chr11_+_20363685
Show fit
3.73
ENST00000530266.5
ENST00000451739.7
ENST00000421577.6
ENST00000443524.6
ENST00000419348.6
HTATIP2
HIV-1 Tat interactive protein 2
chr6_-_41747390
Show fit
3.70
ENST00000356667.8
ENST00000373025.7
ENST00000425343.6
PGC
progastricsin
chr4_-_71784046
Show fit
3.69
ENST00000513476.5
ENST00000273951.13
GC
GC vitamin D binding protein
chr1_-_56966006
Show fit
3.68
ENST00000371237.9
C8B
complement C8 beta chain
chr4_+_73404255
Show fit
3.62
ENST00000621628.4
ENST00000621085.4
ENST00000415165.6
ENST00000295897.9
ENST00000503124.5
ENST00000509063.5
ENST00000401494.7
ALB
albumin
chr20_+_9069076
Show fit
3.60
ENST00000378473.9
PLCB4
phospholipase C beta 4
chr11_-_66345066
Show fit
3.58
ENST00000359957.8
ENST00000425825.6
BRMS1
BRMS1 transcriptional repressor and anoikis regulator
chr8_-_23404076
Show fit
3.52
ENST00000524168.1
ENST00000389131.8
ENST00000523833.2
ENST00000519243.1
LOXL2
lysyl oxidase like 2
chr14_+_51240205
Show fit
3.44
ENST00000457354.7
TMX1
thioredoxin related transmembrane protein 1
chr3_+_130850585
Show fit
3.41
ENST00000505330.5
ENST00000504381.5
ENST00000507488.6
ATP2C1
ATPase secretory pathway Ca2+ transporting 1
chr10_+_113709261
Show fit
3.40
ENST00000672138.1
ENST00000452490.3
CASP7
caspase 7
chr7_+_134745460
Show fit
3.36
ENST00000436461.6
CALD1
caldesmon 1
chr2_-_224569782
Show fit
3.34
ENST00000409096.5
CUL3
cullin 3
chr6_+_125203639
Show fit
3.32
ENST00000392482.6
TPD52L1
TPD52 like 1
chr17_-_28370283
Show fit
3.26
ENST00000226218.9
VTN
vitronectin
chr3_-_197183963
Show fit
3.22
ENST00000653795.1
DLG1
discs large MAGUK scaffold protein 1
chr1_+_74235377
Show fit
3.20
ENST00000326637.8
TNNI3K
TNNI3 interacting kinase
chr4_-_185535498
Show fit
3.18
ENST00000284767.12
ENST00000284771.7
ENST00000284770.10
ENST00000620787.5
ENST00000629667.2
PDLIM3
PDZ and LIM domain 3
chr7_+_77840122
Show fit
3.17
ENST00000450574.5
ENST00000248550.7
PHTF2
putative homeodomain transcription factor 2
chr1_-_246566238
Show fit
3.13
ENST00000366514.5
TFB2M
transcription factor B2, mitochondrial
chr17_+_7583828
Show fit
3.13
ENST00000396501.8
ENST00000250124.11
ENST00000584378.5
ENST00000423172.6
ENST00000579445.5
ENST00000585217.5
ENST00000581380.1
MPDU1
mannose-P-dolichol utilization defect 1
chr14_+_20688756
Show fit
3.06
ENST00000397990.5
ENST00000555597.1
ANG
RNASE4
angiogenin
chr2_+_180981108
Show fit
3.05
ENST00000602710.5
UBE2E3
ubiquitin conjugating enzyme E2 E3
chr17_+_42998379
Show fit
3.01
ENST00000253788.12
ENST00000589913.6
RPL27
ribosomal protein L27
chr3_-_197183806
Show fit
3.00
ENST00000671246.1
ENST00000660553.1
DLG1
discs large MAGUK scaffold protein 1
chr1_-_53940100
Show fit
3.00
ENST00000371376.1
HSPB11
heat shock protein family B (small) member 11
chr10_-_116273606
Show fit
2.96
ENST00000682743.1
GFRA1
GDNF family receptor alpha 1
chr7_+_73828160
Show fit
2.89
ENST00000431918.1
CLDN4
claudin 4
chr8_+_31639755
Show fit
2.86
ENST00000520407.5
NRG1
neuregulin 1
chr10_+_69801874
Show fit
2.86
ENST00000357811.8
COL13A1
collagen type XIII alpha 1 chain
chr1_-_244860376
Show fit
2.85
ENST00000638716.1
HNRNPU
heterogeneous nuclear ribonucleoprotein U
chr12_+_103930600
Show fit
2.85
ENST00000680316.1
ENST00000679861.1
HSP90B1
heat shock protein 90 beta family member 1
chr2_+_28395511
Show fit
2.83
ENST00000436647.1
FOSL2
FOS like 2, AP-1 transcription factor subunit
chr8_+_11809135
Show fit
2.80
ENST00000528643.5
ENST00000525777.5
FDFT1
farnesyl-diphosphate farnesyltransferase 1
chr21_-_37072688
Show fit
2.75
ENST00000464265.5
ENST00000399102.5
PIGP
phosphatidylinositol glycan anchor biosynthesis class P
chr9_-_72953047
Show fit
2.64
ENST00000297785.8
ENST00000376939.5
ALDH1A1
aldehyde dehydrogenase 1 family member A1
chr17_+_7572818
Show fit
2.61
ENST00000293831.13
ENST00000582169.5
ENST00000578754.5
ENST00000578495.5
ENST00000585024.5
ENST00000583802.5
ENST00000577269.5
ENST00000584784.5
ENST00000582746.5
ENST00000581621.1
EIF4A1
ENSG00000264772.6
eukaryotic translation initiation factor 4A1
chr12_-_47705971
Show fit
2.54
ENST00000380650.4
RPAP3
RNA polymerase II associated protein 3
chr22_+_20774092
Show fit
2.52
ENST00000215727.10
SERPIND1
serpin family D member 1
chr4_+_145482761
Show fit
2.52
ENST00000507367.1
ENST00000394092.6
ENST00000515385.1
SMAD1
SMAD family member 1
chr15_-_29822418
Show fit
2.50
ENST00000614355.5
ENST00000495972.6
ENST00000346128.10
TJP1
tight junction protein 1
chr3_-_100114488
Show fit
2.43
ENST00000477258.2
ENST00000354552.7
ENST00000331335.9
ENST00000398326.2
FILIP1L
filamin A interacting protein 1 like
chr3_+_136930469
Show fit
2.43
ENST00000469404.1
ENST00000467911.1
NCK1
NCK adaptor protein 1
chr7_+_28412511
Show fit
2.42
ENST00000357727.7
CREB5
cAMP responsive element binding protein 5
chr7_+_95485898
Show fit
2.37
ENST00000428113.5
ASB4
ankyrin repeat and SOCS box containing 4
chr10_+_122163426
Show fit
2.37
ENST00000360561.7
TACC2
transforming acidic coiled-coil containing protein 2
chr3_-_114179052
Show fit
2.33
ENST00000383673.5
ENST00000295881.9
DRD3
dopamine receptor D3
chr19_+_40751179
Show fit
2.32
ENST00000243563.8
ENST00000601393.1
SNRPA
small nuclear ribonucleoprotein polypeptide A
chr6_-_87095059
Show fit
2.31
ENST00000369582.6
ENST00000610310.3
ENST00000630630.2
ENST00000627148.3
ENST00000625577.1
CGA
glycoprotein hormones, alpha polypeptide
chr10_+_69802424
Show fit
2.30
ENST00000673802.2
ENST00000517713.5
ENST00000520133.5
ENST00000522165.5
ENST00000673641.2
ENST00000673628.2
COL13A1
collagen type XIII alpha 1 chain
chr12_-_47705990
Show fit
2.27
ENST00000432584.7
ENST00000005386.8
RPAP3
RNA polymerase II associated protein 3
chr1_-_246193727
Show fit
2.26
ENST00000391836.3
SMYD3
SET and MYND domain containing 3
chr10_+_122560751
Show fit
2.19
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
DMBT1
deleted in malignant brain tumors 1
chr1_+_152514474
Show fit
2.17
ENST00000368790.4
CRCT1
cysteine rich C-terminal 1
chr9_+_65675834
Show fit
2.14
ENST00000377392.9
ENST00000377384.5
ENST00000430059.6
ENST00000429800.6
ENST00000382405.8
ENST00000377395.8
CBWD5
COBW domain containing 5
chr3_-_197183849
Show fit
2.08
ENST00000443183.5
DLG1
discs large MAGUK scaffold protein 1
chr20_+_11917859
Show fit
2.05
ENST00000618296.4
ENST00000378226.7
BTBD3
BTB domain containing 3
chr2_-_21044063
Show fit
2.04
ENST00000233242.5
APOB
apolipoprotein B
chr20_+_33235987
Show fit
2.03
ENST00000375422.6
ENST00000375413.8
ENST00000354297.9
BPIFA1
BPI fold containing family A member 1
chr10_+_122560679
Show fit
1.98
ENST00000657942.1
DMBT1
deleted in malignant brain tumors 1
chr20_+_37744630
Show fit
1.96
ENST00000373473.5
CTNNBL1
catenin beta like 1
chr10_+_122560639
Show fit
1.96
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
DMBT1
deleted in malignant brain tumors 1
chr8_-_94436926
Show fit
1.94
ENST00000481490.3
FSBP
fibrinogen silencer binding protein
chr2_-_86195400
Show fit
1.92
ENST00000442664.6
ENST00000409051.6
ENST00000410111.8
ENST00000620815.4
ENST00000449247.6
IMMT
inner membrane mitochondrial protein
chr17_+_74431338
Show fit
1.90
ENST00000342648.9
ENST00000652232.1
ENST00000481232.2
GPRC5C
G protein-coupled receptor class C group 5 member C
chr4_+_168092530
Show fit
1.89
ENST00000359299.8
ANXA10
annexin A10
chr3_+_137998735
Show fit
1.88
ENST00000343735.8
CLDN18
claudin 18
chr1_+_23691742
Show fit
1.88
ENST00000374550.8
ENST00000643754.2
RPL11
ribosomal protein L11
chr8_-_80080816
Show fit
1.82
ENST00000520527.5
ENST00000517427.5
ENST00000379097.7
ENST00000448733.3
TPD52
tumor protein D52
chr12_+_21372899
Show fit
1.82
ENST00000240652.8
ENST00000542023.1
ENST00000537593.1
IAPP
islet amyloid polypeptide
chr17_+_76376581
Show fit
1.80
ENST00000591651.5
ENST00000545180.5
SPHK1
sphingosine kinase 1
chr7_+_117014881
Show fit
1.72
ENST00000422922.5
ENST00000432298.5
ST7
suppression of tumorigenicity 7
chr1_+_207104226
Show fit
1.72
ENST00000367070.8
C4BPA
complement component 4 binding protein alpha
chr22_-_21227637
Show fit
1.69
ENST00000401924.5
GGT2
gamma-glutamyltransferase 2
chr3_+_69936629
Show fit
1.69
ENST00000394348.2
ENST00000531774.1
MITF
melanocyte inducing transcription factor
chr4_-_102345469
Show fit
1.69
ENST00000356736.5
ENST00000682932.1
SLC39A8
solute carrier family 39 member 8
chr6_+_5260992
Show fit
1.64
ENST00000324331.10
FARS2
phenylalanyl-tRNA synthetase 2, mitochondrial
chr3_+_14178808
Show fit
1.63
ENST00000306024.4
LSM3
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated
chr15_+_49170200
Show fit
1.63
ENST00000396509.6
GALK2
galactokinase 2
chr7_+_130266847
Show fit
1.62
ENST00000222481.9
CPA2
carboxypeptidase A2
chr1_-_167553745
Show fit
1.60
ENST00000370509.5
CREG1
cellular repressor of E1A stimulated genes 1
chr20_-_7940444
Show fit
1.58
ENST00000378789.4
HAO1
hydroxyacid oxidase 1
chr14_-_25010604
Show fit
1.56
ENST00000550887.5
STXBP6
syntaxin binding protein 6
chr11_-_6440980
Show fit
1.50
ENST00000265983.8
ENST00000615166.1
HPX
hemopexin
chr5_+_138338256
Show fit
1.50
ENST00000513056.5
ENST00000239906.10
ENST00000511276.1
FAM53C
family with sequence similarity 53 member C
chr4_-_185775271
Show fit
1.47
ENST00000430503.5
ENST00000319454.10
ENST00000450341.5
SORBS2
sorbin and SH3 domain containing 2
chr2_-_46617020
Show fit
1.45
ENST00000474980.1
ENST00000281382.11
ENST00000306465.8
PIGF
phosphatidylinositol glycan anchor biosynthesis class F
chr8_+_19313685
Show fit
1.44
ENST00000265807.8
ENST00000518040.5
SH2D4A
SH2 domain containing 4A
chr1_-_169586471
Show fit
1.43
ENST00000367797.9
F5
coagulation factor V
chr14_+_20343607
Show fit
1.42
ENST00000250416.9
ENST00000429687.8
ENST00000527915.5
PARP2
poly(ADP-ribose) polymerase 2
chr17_+_9021501
Show fit
1.42
ENST00000173229.7
NTN1
netrin 1
chr9_-_92424427
Show fit
1.42
ENST00000375550.5
OMD
osteomodulin
chr10_+_103555124
Show fit
1.40
ENST00000437579.1
NEURL1
neuralized E3 ubiquitin protein ligase 1
chr15_-_59372863
Show fit
1.39
ENST00000288235.9
MYO1E
myosin IE
chr5_+_143812161
Show fit
1.36
ENST00000289448.4
HMHB1
histocompatibility minor HB-1
chr11_-_115504389
Show fit
1.34
ENST00000545380.5
ENST00000452722.7
ENST00000331581.11
ENST00000537058.5
ENST00000536727.5
ENST00000542447.6
CADM1
cell adhesion molecule 1
chr4_-_69495897
Show fit
1.34
ENST00000305107.7
ENST00000639621.1
UGT2B4
UDP glucuronosyltransferase family 2 member B4
chrX_-_134658450
Show fit
1.33
ENST00000359237.9
PLAC1
placenta enriched 1
chr21_-_42366525
Show fit
1.31
ENST00000291527.3
TFF1
trefoil factor 1
chr3_+_155870623
Show fit
1.28
ENST00000295920.7
ENST00000496455.7
GMPS
guanine monophosphate synthase
chr3_+_136022734
Show fit
1.28
ENST00000334546.6
PPP2R3A
protein phosphatase 2 regulatory subunit B''alpha
chr4_-_69495861
Show fit
1.22
ENST00000512583.5
UGT2B4
UDP glucuronosyltransferase family 2 member B4
chr4_-_122456725
Show fit
1.22
ENST00000226730.5
IL2
interleukin 2
chr15_-_66504832
Show fit
1.19
ENST00000569438.2
ENST00000569696.5
ENST00000307961.11
RPL4
ribosomal protein L4
chr9_-_107489754
Show fit
1.17
ENST00000610832.1
ENST00000374672.5
KLF4
Kruppel like factor 4
chr12_+_57694118
Show fit
1.16
ENST00000315970.12
ENST00000547079.5
ENST00000439210.6
ENST00000389146.10
ENST00000413095.6
ENST00000551035.5
ENST00000257966.12
ENST00000435406.6
ENST00000550372.5
ENST00000389142.9
OS9
OS9 endoplasmic reticulum lectin
chr6_+_10528326
Show fit
1.15
ENST00000379597.7
GCNT2
glucosaminyl (N-acetyl) transferase 2 (I blood group)
chr1_-_43389768
Show fit
1.14
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
MED8
mediator complex subunit 8
chrX_+_136532205
Show fit
1.12
ENST00000370634.8
VGLL1
vestigial like family member 1
chr10_+_35195124
Show fit
1.12
ENST00000487763.5
ENST00000473940.5
ENST00000488328.5
ENST00000356917.9
CREM
cAMP responsive element modulator
chr5_-_41213505
Show fit
1.10
ENST00000337836.10
ENST00000433294.1
C6
complement C6
chr17_-_352784
Show fit
1.09
ENST00000577079.5
ENST00000331302.12
ENST00000618002.4
ENST00000536489.6
RPH3AL
rabphilin 3A like (without C2 domains)
chr6_-_112254647
Show fit
1.06
ENST00000455073.1
ENST00000522006.5
ENST00000519932.5
LAMA4
laminin subunit alpha 4
chr6_-_25830557
Show fit
1.06
ENST00000468082.1
SLC17A1
solute carrier family 17 member 1
chr15_+_49170237
Show fit
1.02
ENST00000560031.6
ENST00000558145.5
ENST00000544523.5
ENST00000560138.5
GALK2
galactokinase 2
chr3_+_138010143
Show fit
1.01
ENST00000183605.10
CLDN18
claudin 18
chr15_+_75043263
Show fit
1.01
ENST00000563393.1
PPCDC
phosphopantothenoylcysteine decarboxylase
chr5_+_148394712
Show fit
1.00
ENST00000513826.1
FBXO38
F-box protein 38
chr3_+_159852933
Show fit
0.98
ENST00000482804.1
SCHIP1
schwannomin interacting protein 1
chr12_-_102478539
Show fit
0.97
ENST00000424202.6
IGF1
insulin like growth factor 1
chr13_+_52024691
Show fit
0.97
ENST00000521776.2
UTP14C
UTP14C small subunit processome component
chr4_-_169757873
Show fit
0.94
ENST00000393381.3
HPF1
histone PARylation factor 1
chr12_-_14885845
Show fit
0.93
ENST00000539261.6
ENST00000228938.5
MGP
matrix Gla protein
chr4_-_115113614
Show fit
0.91
ENST00000264363.7
NDST4
N-deacetylase and N-sulfotransferase 4
chr6_-_24666591
Show fit
0.91
ENST00000341060.3
TDP2
tyrosyl-DNA phosphodiesterase 2
chr12_-_71157872
Show fit
0.89
ENST00000546561.2
TSPAN8
tetraspanin 8
chr13_-_74133892
Show fit
0.89
ENST00000377669.7
KLF12
Kruppel like factor 12
chr1_-_28058087
Show fit
0.89
ENST00000373864.5
EYA3
EYA transcriptional coactivator and phosphatase 3
chr1_+_179882275
Show fit
0.88
ENST00000606911.7
ENST00000271583.7
TOR1AIP1
torsin 1A interacting protein 1
chr3_+_148730100
Show fit
0.88
ENST00000474935.5
ENST00000475347.5
ENST00000461609.1
AGTR1
angiotensin II receptor type 1
chr7_-_25228485
Show fit
0.86
ENST00000222674.2
NPVF
neuropeptide VF precursor
chr6_+_26251607
Show fit
0.85
ENST00000619466.2
H2BC9
H2B clustered histone 9
chr19_-_45584769
Show fit
0.84
ENST00000263275.5
OPA3
outer mitochondrial membrane lipid metabolism regulator OPA3
chr19_+_926001
Show fit
0.84
ENST00000263620.8
ARID3A
AT-rich interaction domain 3A
chr1_-_169586539
Show fit
0.83
ENST00000367796.3
F5
coagulation factor V
chr9_-_14722725
Show fit
0.83
ENST00000380911.4
CER1
cerberus 1, DAN family BMP antagonist
chr18_-_3874247
Show fit
0.82
ENST00000581699.5
DLGAP1
DLG associated protein 1