Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
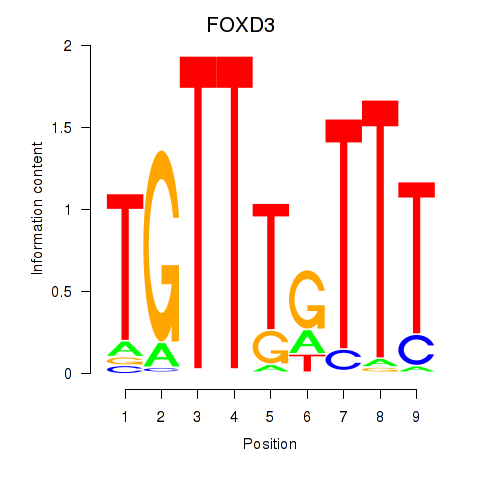
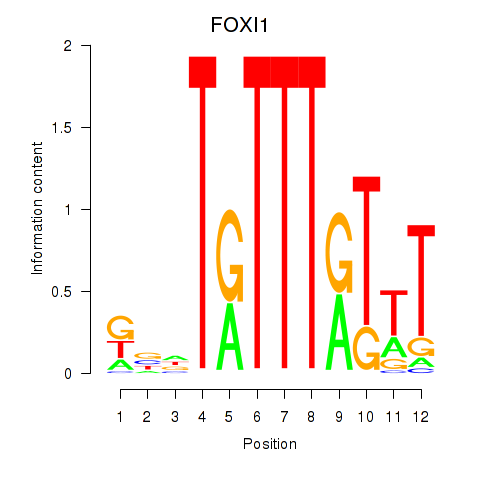
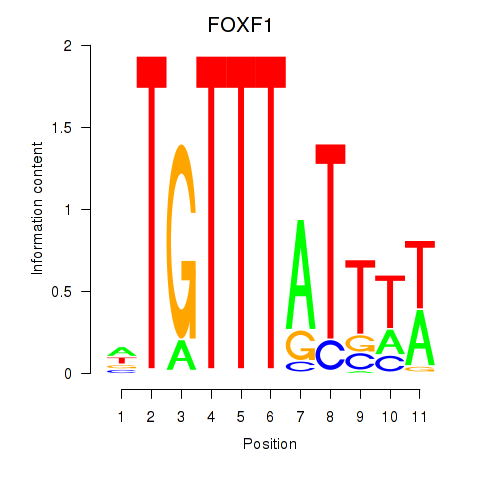
Results for FOXD3_FOXI1_FOXF1
Z-value: 1.01
Transcription factors associated with FOXD3_FOXI1_FOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD3
|
ENSG00000187140.6 | FOXD3 |
|
FOXI1
|
ENSG00000168269.10 | FOXI1 |
|
FOXF1
|
ENSG00000103241.7 | FOXF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXI1 | hg38_v1_chr5_+_170105892_170105913 | 0.21 | 2.0e-03 | Click! |
| FOXF1 | hg38_v1_chr16_+_86510507_86510539 | 0.18 | 6.8e-03 | Click! |
| FOXD3 | hg38_v1_chr1_+_63322558_63322583 | 0.06 | 3.6e-01 | Click! |
Activity profile of FOXD3_FOXI1_FOXF1 motif
Sorted Z-values of FOXD3_FOXI1_FOXF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD3_FOXI1_FOXF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 4.4 | 17.6 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 4.3 | 17.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 3.9 | 66.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 3.8 | 15.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.3 | 9.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 3.3 | 19.5 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 3.1 | 24.8 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 3.0 | 24.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 2.1 | 8.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.6 | 19.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 20.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 1.3 | 5.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.3 | 11.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.2 | 8.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.2 | 3.6 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.2 | 4.7 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.2 | 5.8 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 1.1 | 13.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 1.1 | 3.4 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 1.1 | 11.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.1 | 12.7 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 1.0 | 3.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 1.0 | 16.5 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 1.0 | 16.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.0 | 6.0 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 1.0 | 3.8 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.9 | 12.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.8 | 12.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.8 | 4.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.8 | 4.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.8 | 12.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.7 | 2.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.7 | 2.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.7 | 2.8 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.7 | 2.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.7 | 1.3 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.7 | 2.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.7 | 3.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.6 | 3.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 0.6 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.6 | 2.5 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 3.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.6 | 1.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 1.8 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.6 | 1.8 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.6 | 2.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 4.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 2.9 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.6 | 4.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.6 | 7.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.6 | 8.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.6 | 5.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.6 | 3.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.5 | 1.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 3.8 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.5 | 4.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.5 | 1.6 | GO:0072425 | cellular response to nitrosative stress(GO:0071500) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 | 2.7 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.5 | 2.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.5 | 2.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 1.0 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.5 | 3.6 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.5 | 2.5 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.5 | 38.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.5 | 4.4 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.5 | 3.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.5 | 12.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 1.4 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.4 | 1.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 6.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 1.7 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.4 | 1.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 1.2 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 3.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 3.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 3.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.4 | 5.2 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.4 | 1.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.4 | 0.8 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.4 | 1.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 2.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 7.6 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.4 | 1.4 | GO:0072011 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 0.4 | 1.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.4 | 1.1 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.4 | 2.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 10.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 3.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.3 | 3.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 7.4 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.3 | 4.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 | 5.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 5.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 3.6 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.3 | 3.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.3 | 15.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.3 | 1.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 3.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 1.3 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 2.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) Golgi disassembly(GO:0090166) |
| 0.3 | 29.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.3 | 0.9 | GO:0090135 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) actin filament branching(GO:0090135) |
| 0.3 | 5.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 11.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.3 | 0.6 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.3 | 3.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 1.2 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 12.2 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.3 | 2.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 2.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 10.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.3 | 14.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.3 | 7.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 | 1.1 | GO:0072007 | mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.3 | 0.8 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.3 | 1.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.3 | 2.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 0.8 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 1.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 1.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 1.9 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 0.9 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 1.4 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.2 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 2.4 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.2 | 1.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 4.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.8 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 1.9 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 0.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.6 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 1.5 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.2 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 1.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 0.8 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 | 0.6 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.8 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 0.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 0.8 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.2 | 0.8 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.2 | 8.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 3.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 0.4 | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 0.2 | 3.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.2 | 0.6 | GO:0033168 | regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.2 | 2.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 1.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.8 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 | 0.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 | 0.5 | GO:1990910 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.2 | 2.7 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.2 | 3.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 3.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 1.2 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.2 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.5 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.2 | 0.8 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.2 | 0.5 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 3.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 0.7 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 2.8 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 1.8 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.8 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 1.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 1.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 0.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 6.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.2 | 4.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 1.4 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.2 | 4.7 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.2 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 4.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 2.0 | GO:1990440 | ATF6-mediated unfolded protein response(GO:0036500) positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 1.8 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 1.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 | 1.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 23.8 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 0.6 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.1 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 2.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 1.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.3 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.8 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 1.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 3.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.4 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.1 | 4.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.7 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.6 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 0.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.9 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 2.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.7 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 0.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 3.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.1 | GO:0038001 | paracrine signaling(GO:0038001) |
| 0.1 | 0.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.4 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 1.9 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 0.6 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.1 | 0.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 7.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.3 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 1.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.6 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.6 | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.1 | 1.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 2.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 1.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 2.2 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.7 | GO:0015865 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 2.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.7 | GO:1901740 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) positive regulation of hepatocyte proliferation(GO:2000347) |
| 0.1 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 2.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 3.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 14.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.6 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 3.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 2.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 1.4 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 7.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.3 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.1 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.3 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.3 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 1.1 | GO:0090314 | positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.2 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 1.0 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 2.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 2.2 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.5 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 2.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0072540 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 1.2 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:1901748 | glutathione catabolic process(GO:0006751) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.8 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.4 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.2 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) C-5 methylation of cytosine(GO:0090116) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 1.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.0 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0032747 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.9 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.8 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.5 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.3 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 67.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.6 | 17.9 | GO:0072534 | perineuronal net(GO:0072534) |
| 2.5 | 17.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 2.0 | 9.8 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.7 | 19.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.6 | 32.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.0 | 3.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.0 | 4.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.9 | 11.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.6 | 25.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.5 | 2.7 | GO:0001652 | granular component(GO:0001652) |
| 0.5 | 6.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 3.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) dendritic spine neck(GO:0044326) |
| 0.5 | 3.3 | GO:0019867 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.4 | 1.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 1.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.4 | 5.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 13.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.4 | 4.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 1.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 1.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 5.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 2.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 11.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 1.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 3.3 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.3 | 0.8 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 0.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 6.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 3.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.7 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 4.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 7.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 4.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 0.6 | GO:0033167 | ARC complex(GO:0033167) |
| 0.2 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 2.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 4.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 5.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 31.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 1.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 11.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.5 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.2 | 0.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 6.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 0.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 2.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 2.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.8 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 32.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.7 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 13.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 11.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 29.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 4.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 4.3 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 15.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 1.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 15.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 3.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 25.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 2.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 5.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 5.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 35.7 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 2.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 2.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 5.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.0 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) intrinsic component of organelle membrane(GO:0031300) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 69.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.3 | 13.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 2.3 | 67.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.2 | 17.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 2.1 | 8.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 2.0 | 17.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.0 | 9.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.8 | 27.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.5 | 4.6 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.5 | 25.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.5 | 18.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.5 | 16.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.4 | 4.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.4 | 5.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.3 | 3.8 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.2 | 25.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.2 | 3.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.1 | 3.2 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 1.1 | 3.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.1 | 12.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 11.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.7 | 24.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.7 | 11.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.7 | 3.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 10.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 1.9 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.6 | 13.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.6 | 2.5 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.6 | 28.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.6 | 9.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.6 | 5.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 18.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 3.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.6 | 2.8 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.6 | 7.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 2.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.5 | 4.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.5 | 22.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 2.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.5 | 8.5 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.5 | 1.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.5 | 2.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.5 | 3.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 1.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.5 | 6.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 5.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 4.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.5 | 24.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 2.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 1.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 2.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 3.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 1.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.4 | 2.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.4 | 2.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.4 | 11.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 2.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.4 | 1.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 2.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.4 | 5.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 2.6 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.4 | 2.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 0.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.4 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 27.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 3.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 1.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 1.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 3.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 0.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 3.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 1.8 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 1.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 1.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.3 | 0.8 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.3 | 2.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 3.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 5.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 8.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 3.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 0.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 13.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 1.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.2 | 3.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.2 | 2.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 6.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 2.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 2.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 2.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 0.8 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 1.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 1.8 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.2 | 0.9 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.2 | 2.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 3.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 1.9 | GO:0042577 | phosphatidate phosphatase activity(GO:0008195) lipid phosphatase activity(GO:0042577) |
| 0.2 | 2.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 8.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 0.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 2.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.8 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 8.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.8 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.1 | 1.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 4.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 2.0 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 2.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 4.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 3.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 3.4 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 8.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 37.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 3.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 4.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.8 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.6 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 2.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 9.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 4.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.3 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 6.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 6.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 5.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.6 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 4.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 12.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 26.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 25.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 19.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 3.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 15.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 12.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 1.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 45.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 9.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 13.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 9.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 14.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 11.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 3.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 31.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.7 | 1.5 | REACTOME TRIF MEDIATED TLR3 SIGNALING | Genes involved in TRIF mediated TLR3 signaling |
| 0.7 | 49.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.7 | 13.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.6 | 14.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.6 | 24.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 10.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 9.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 16.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 9.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 12.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 12.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 3.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 10.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 5.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 6.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 5.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 10.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.9 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 6.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 2.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 5.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 4.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 16.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.1 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 5.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 6.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 3.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.2 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 5.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |