Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
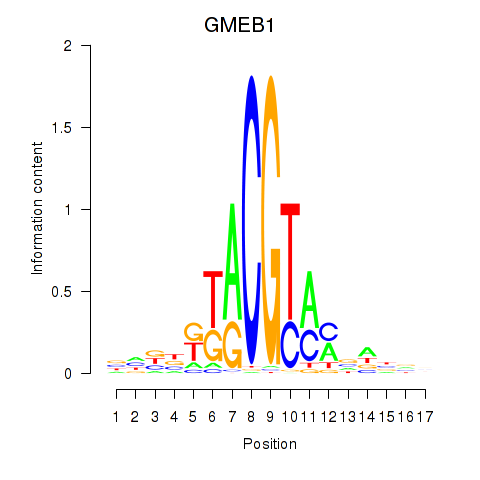
Results for GMEB1
Z-value: 1.04
Transcription factors associated with GMEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GMEB1
|
ENSG00000162419.13 | GMEB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB1 | hg38_v1_chr1_+_28668746_28668815 | 0.22 | 8.6e-04 | Click! |
Activity profile of GMEB1 motif
Sorted Z-values of GMEB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GMEB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 21.2 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 3.6 | 10.9 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 3.1 | 12.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 3.0 | 24.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 2.7 | 10.7 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 2.4 | 9.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 2.0 | 6.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 1.9 | 9.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.8 | 5.5 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 1.8 | 10.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.8 | 7.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.7 | 8.7 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 1.7 | 23.5 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 1.6 | 8.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 1.6 | 4.7 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 1.6 | 4.7 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.6 | 14.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.4 | 5.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.4 | 6.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 1.3 | 7.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.2 | 3.7 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 1.2 | 3.7 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.2 | 7.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.2 | 4.7 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 1.2 | 3.5 | GO:0036233 | glycine import(GO:0036233) |
| 1.2 | 12.9 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.2 | 5.8 | GO:0015862 | uridine transport(GO:0015862) |
| 1.2 | 5.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.2 | 6.9 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.1 | 3.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.1 | 3.4 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 1.1 | 10.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.1 | 8.9 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 1.1 | 3.2 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 1.1 | 7.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 1.1 | 3.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 1.1 | 3.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 1.0 | 1.0 | GO:1904882 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 1.0 | 6.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.0 | 14.4 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 1.0 | 6.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.0 | 3.0 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 1.0 | 3.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 1.0 | 2.0 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.9 | 8.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.9 | 2.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.9 | 2.7 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.9 | 11.9 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.9 | 3.5 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.9 | 1.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.9 | 4.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.9 | 2.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.8 | 3.4 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.8 | 3.3 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.8 | 2.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.8 | 3.2 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.8 | 1.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.8 | 2.4 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 0.8 | 5.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.7 | 2.2 | GO:1904387 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.7 | 4.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.7 | 0.7 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.7 | 7.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.7 | 2.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.7 | 2.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.7 | 3.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.7 | 2.1 | GO:0042323 | germinal center B cell differentiation(GO:0002314) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) xanthine metabolic process(GO:0046110) deoxyribonucleoside catabolic process(GO:0046121) negative regulation of mucus secretion(GO:0070256) |
| 0.7 | 9.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.7 | 5.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.7 | 2.7 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.7 | 3.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.7 | 2.7 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.7 | 4.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.7 | 2.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.7 | 2.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.7 | 2.6 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.6 | 2.6 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.6 | 3.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.6 | 1.9 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.6 | 4.9 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.6 | 1.8 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.6 | 1.8 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.6 | 1.8 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.6 | 1.8 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.6 | 1.2 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.6 | 1.7 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.6 | 1.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.6 | 2.8 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.5 | 1.6 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.5 | 0.5 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.5 | 3.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.5 | 1.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.5 | 1.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 11.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.5 | 1.0 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.5 | 1.5 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.5 | 1.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.5 | 2.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.5 | 0.5 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.5 | 7.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.5 | 3.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.5 | 1.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.5 | 1.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 19.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.5 | 5.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.5 | 2.8 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.5 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 3.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 2.7 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.4 | 4.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 15.0 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.4 | 2.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.4 | 2.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.4 | 3.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 | 1.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.4 | 2.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 7.7 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.4 | 1.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.4 | 10.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 6.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.4 | 6.8 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.4 | 2.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 2.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.4 | 3.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.4 | 1.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.4 | 1.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.4 | 1.8 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.4 | 1.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 2.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.3 | 2.8 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 8.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 22.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 2.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.3 | 36.9 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.3 | 1.0 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.3 | 1.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.3 | 1.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 1.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 1.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 1.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.0 | GO:0061074 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 2.6 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.3 | 1.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.3 | 6.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 0.9 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.3 | 3.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 0.9 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 1.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.3 | 3.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.3 | 3.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 1.2 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.3 | 2.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.3 | 3.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 0.9 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.3 | 0.6 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 0.9 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 2.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 21.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 5.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.3 | 1.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 9.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 2.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.3 | 1.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 2.4 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.3 | 0.8 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 1.0 | GO:0009838 | abscission(GO:0009838) |
| 0.3 | 2.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 0.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.3 | 1.8 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.3 | 0.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 1.5 | GO:0015853 | adenine transport(GO:0015853) |
| 0.3 | 2.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 3.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 4.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 1.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.2 | 1.7 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.2 | 1.0 | GO:0051586 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 0.7 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 4.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 8.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 2.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 2.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.7 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) regulation of late endosome to lysosome transport(GO:1902822) |
| 0.2 | 0.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 1.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 | 1.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 1.8 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.2 | 0.2 | GO:1990592 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 2.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 3.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 0.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 0.6 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 2.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 8.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 1.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 28.1 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.2 | 7.0 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 0.6 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 3.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 6.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 2.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 1.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 4.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 1.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 0.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.3 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.6 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.2 | 36.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.2 | 0.7 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.2 | 1.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 1.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 5.0 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.3 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.2 | 4.4 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 0.3 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 3.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 2.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 2.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 0.6 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 1.3 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.2 | 2.2 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.2 | 2.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.2 | 2.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 1.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 14.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.2 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 3.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.2 | 0.8 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.2 | 1.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.2 | 0.6 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 6.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.2 | GO:0006228 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.7 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 2.3 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 2.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 2.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 7.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 2.7 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 1.2 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 2.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 3.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.5 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.1 | 0.5 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.7 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 2.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 1.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 2.0 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 1.0 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 0.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.9 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.5 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 1.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 2.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.4 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.1 | 0.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 2.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.5 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 3.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.3 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.7 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 2.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.3 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.4 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 1.0 | GO:0010452 | histone H3-K36 methylation(GO:0010452) peptidyl-lysine monomethylation(GO:0018026) peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 2.5 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.1 | 0.1 | GO:0071888 | macrophage apoptotic process(GO:0071888) |
| 0.1 | 0.2 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.1 | 0.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.8 | GO:0006999 | nuclear pore organization(GO:0006999) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.5 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 5.1 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 2.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 4.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.8 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.0 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.8 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.3 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 1.6 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 3.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.8 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 2.8 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 1.6 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.5 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 1.3 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.4 | GO:0039536 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 1.1 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 4.4 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.8 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 1.1 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.4 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 2.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.8 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.3 | GO:0055026 | negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.0 | 0.8 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0014856 | skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.3 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 12.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 2.3 | 9.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.9 | 5.7 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 1.5 | 6.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.5 | 4.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 1.5 | 13.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 1.5 | 4.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.4 | 26.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 1.2 | 3.7 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.2 | 13.8 | GO:0071203 | WASH complex(GO:0071203) |
| 1.1 | 13.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.1 | 8.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.9 | 11.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.9 | 4.6 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.9 | 2.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.9 | 3.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.9 | 4.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.9 | 6.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.9 | 2.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.8 | 3.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.8 | 8.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 3.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.7 | 3.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.7 | 2.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.7 | 4.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.7 | 2.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.7 | 2.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.7 | 3.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.7 | 13.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.7 | 18.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.7 | 2.0 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.7 | 7.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.7 | 9.9 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.6 | 3.8 | GO:0002177 | manchette(GO:0002177) |
| 0.6 | 3.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.6 | 1.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.6 | 3.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.6 | 18.8 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.6 | 4.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 2.2 | GO:0035061 | Smc5-Smc6 complex(GO:0030915) interchromatin granule(GO:0035061) |
| 0.6 | 2.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.6 | 5.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.6 | 2.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 7.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 1.6 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.5 | 1.6 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.5 | 1.6 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.5 | 2.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.5 | 2.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.5 | 2.5 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.5 | 25.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.5 | 2.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.5 | 8.3 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.5 | 5.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.5 | 5.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 2.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 3.0 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 2.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.4 | 1.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.4 | 4.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.4 | 1.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.4 | 1.9 | GO:0001652 | granular component(GO:0001652) |
| 0.4 | 3.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.4 | 2.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.4 | 4.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.3 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 5.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 7.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 3.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 5.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 1.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 2.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 0.8 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.3 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 2.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 6.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 38.4 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.3 | 0.8 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 2.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 4.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.5 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 1.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 4.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 0.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 1.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 2.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 4.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 2.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 2.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 1.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 5.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 0.8 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.2 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 3.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.8 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 2.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 2.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 4.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 2.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 2.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 1.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 1.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 4.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 8.1 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 4.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 3.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 10.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 4.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 0.6 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.2 | 0.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 2.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 9.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 17.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 2.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 13.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.6 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 3.6 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.5 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.1 | 11.9 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 3.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 31.5 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.8 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 8.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.7 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 4.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 3.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 26.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.5 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 5.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 6.4 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 5.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 36.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0030880 | DNA-directed RNA polymerase complex(GO:0000428) RNA polymerase complex(GO:0030880) nuclear DNA-directed RNA polymerase complex(GO:0055029) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 2.7 | 10.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.6 | 10.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 2.3 | 9.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 2.3 | 6.8 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 2.2 | 28.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.8 | 17.9 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.8 | 7.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.8 | 10.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 1.7 | 5.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.5 | 13.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.5 | 23.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.4 | 8.5 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 1.4 | 5.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.4 | 4.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 1.3 | 3.9 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 1.3 | 7.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 1.3 | 5.0 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 1.2 | 3.5 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 1.1 | 3.4 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 1.1 | 3.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.9 | 5.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.9 | 2.7 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.9 | 2.6 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.8 | 7.4 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.8 | 2.4 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.8 | 2.4 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.8 | 3.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.7 | 2.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.7 | 2.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.7 | 1.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.7 | 2.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.7 | 2.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 2.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.7 | 4.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.7 | 2.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.7 | 2.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.7 | 2.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.6 | 3.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.6 | 1.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.6 | 2.3 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.6 | 2.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.6 | 3.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 2.8 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.5 | 2.2 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.5 | 1.6 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.5 | 2.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 3.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.5 | 4.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.5 | 4.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.5 | 17.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 3.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.5 | 4.5 | GO:0043184 | morphogen activity(GO:0016015) vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 2.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.5 | 5.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.5 | 1.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.5 | 1.9 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.5 | 1.9 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.5 | 2.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 5.9 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.4 | 4.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.4 | 5.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 3.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 12.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 1.7 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.4 | 9.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.4 | 3.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 4.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.4 | 3.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 5.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 1.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.4 | 2.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 1.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 2.7 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 3.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.4 | 1.9 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 4.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.4 | 11.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 2.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.4 | 3.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 6.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 10.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 3.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 6.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 12.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.3 | 1.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.3 | 1.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 4.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 2.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 0.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.3 | 4.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 2.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 3.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 5.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 3.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.3 | 1.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 1.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.3 | 1.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 0.9 | GO:0000035 | acyl binding(GO:0000035) |
| 0.3 | 2.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 0.9 | GO:0051717 | phosphohistidine phosphatase activity(GO:0008969) bisphosphoglycerate phosphatase activity(GO:0034416) inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.3 | 2.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 0.9 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.3 | 8.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 2.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 2.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 2.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 0.9 | GO:0016154 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.3 | 6.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 0.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.3 | 37.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 1.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 17.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 0.8 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 0.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 0.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 3.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 1.8 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 2.7 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.2 | 3.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 2.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 5.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 2.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 2.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 5.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 3.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 0.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 6.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 3.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 4.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 5.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 2.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 3.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 3.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 13.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 2.7 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.2 | 3.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 9.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 10.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.2 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.8 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 0.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 0.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 2.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 0.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.8 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 10.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 3.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 2.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 27.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 1.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 1.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 2.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 3.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.7 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 4.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 2.8 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 2.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 0.1 | 2.0 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 1.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 6.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.9 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 2.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 3.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 8.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 4.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 3.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 1.6 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 3.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 7.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 5.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 3.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 30.8 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 13.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 25.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 11.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 20.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 27.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 7.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 12.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 12.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 12.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 7.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 1.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 8.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 6.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 3.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 9.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 4.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 4.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 6.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 3.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 3.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 6.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 20.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.3 | 30.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.2 | 35.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.8 | 12.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.8 | 13.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.7 | 23.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.7 | 7.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.6 | 17.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.6 | 20.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.6 | 4.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.5 | 2.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 11.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 35.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.5 | 10.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.4 | 6.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 5.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 7.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 49.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.3 | 3.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 7.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.3 | 6.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 35.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 5.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.2 | 4.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 2.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 0.9 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 4.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 0.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 5.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 7.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 4.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 7.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 0.6 | REACTOME NUCLEOTIDE EXCISION REPAIR | Genes involved in Nucleotide Excision Repair |
| 0.2 | 1.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 4.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 4.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 7.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 4.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 9.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 8.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 4.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 8.8 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 2.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 2.2 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 4.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.7 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.6 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 2.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |