Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HIC1
Z-value: 2.13
Transcription factors associated with HIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIC1
|
ENSG00000177374.13 | HIC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIC1 | hg38_v1_chr17_+_2056073_2056310, hg38_v1_chr17_+_2055094_2055110 | 0.46 | 1.1e-12 | Click! |
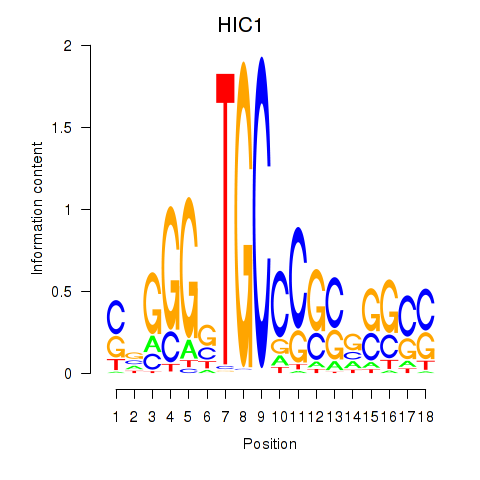
Activity profile of HIC1 motif
Sorted Z-values of HIC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.3 | 65.0 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 11.2 | 67.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 9.8 | 29.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 8.8 | 26.4 | GO:0070079 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 8.4 | 25.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 7.6 | 22.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 7.6 | 30.3 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 7.1 | 21.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 6.8 | 34.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 6.8 | 54.5 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 6.6 | 33.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 6.4 | 25.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 6.3 | 31.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 6.0 | 17.9 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 5.9 | 5.9 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 5.8 | 17.5 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 5.8 | 29.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 5.5 | 65.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 5.3 | 10.5 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 5.3 | 26.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 5.2 | 31.4 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 4.9 | 19.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 4.6 | 9.2 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 4.6 | 91.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 4.5 | 36.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 4.4 | 22.2 | GO:1904207 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 4.4 | 17.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 4.3 | 21.7 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 4.3 | 17.0 | GO:1904021 | regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 4.2 | 25.3 | GO:0015853 | adenine transport(GO:0015853) |
| 4.2 | 12.6 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 4.2 | 12.5 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 4.1 | 20.6 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 4.0 | 16.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 4.0 | 16.0 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 4.0 | 15.9 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 4.0 | 15.9 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 3.9 | 11.7 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 3.9 | 11.7 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 3.8 | 7.6 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 3.8 | 7.6 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 3.8 | 22.7 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 3.7 | 11.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 3.5 | 10.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 3.3 | 16.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 3.2 | 3.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 3.2 | 9.6 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 3.2 | 9.5 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 3.2 | 12.6 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 3.1 | 28.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 3.1 | 9.4 | GO:0019075 | virus maturation(GO:0019075) |
| 3.1 | 9.3 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 3.1 | 24.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 3.1 | 9.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 3.0 | 57.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.9 | 11.8 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 2.9 | 14.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 2.9 | 14.4 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 2.8 | 5.7 | GO:1902722 | positive regulation of prolactin secretion(GO:1902722) |
| 2.8 | 11.3 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 2.8 | 16.7 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 2.8 | 49.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 2.7 | 19.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 2.7 | 35.4 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 2.6 | 7.9 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 2.6 | 15.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 2.6 | 18.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 2.6 | 30.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 2.5 | 22.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 2.4 | 7.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 2.4 | 19.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 2.4 | 21.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 2.4 | 21.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 2.4 | 7.1 | GO:0022012 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 2.3 | 18.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.3 | 9.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 2.3 | 11.5 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 2.3 | 29.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 2.3 | 9.0 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 2.2 | 13.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 2.2 | 6.7 | GO:0001757 | somite specification(GO:0001757) |
| 2.2 | 13.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.2 | 6.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 2.2 | 6.5 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 2.2 | 6.5 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 2.2 | 64.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 2.2 | 17.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 2.1 | 6.4 | GO:0007343 | egg activation(GO:0007343) |
| 2.1 | 8.4 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 2.1 | 6.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 2.1 | 10.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 2.1 | 14.5 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 2.0 | 12.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 2.0 | 16.0 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 2.0 | 9.9 | GO:0015808 | L-alanine transport(GO:0015808) |
| 2.0 | 9.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 2.0 | 11.8 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.9 | 3.9 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 1.9 | 15.5 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 1.9 | 13.6 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.9 | 13.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 1.9 | 5.8 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.9 | 11.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.9 | 36.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 1.9 | 7.6 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.9 | 7.5 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 1.9 | 9.3 | GO:0051029 | rRNA transport(GO:0051029) |
| 1.8 | 5.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.8 | 1.8 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 1.7 | 13.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.7 | 6.9 | GO:0060437 | lung growth(GO:0060437) |
| 1.7 | 12.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.7 | 17.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.7 | 8.6 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 1.7 | 5.2 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 1.7 | 8.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.7 | 6.8 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 1.7 | 13.5 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 1.6 | 14.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 1.6 | 6.5 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 1.6 | 6.5 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 1.6 | 11.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 1.6 | 3.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 1.6 | 1.6 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 1.6 | 54.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.6 | 9.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 1.6 | 15.6 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 1.6 | 4.7 | GO:1901859 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 1.5 | 7.7 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 1.5 | 6.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.5 | 35.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 1.5 | 7.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.5 | 7.7 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 1.5 | 7.6 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.5 | 4.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 1.5 | 10.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 1.5 | 8.8 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.5 | 8.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.4 | 4.3 | GO:0072177 | mesonephric duct development(GO:0072177) |
| 1.4 | 8.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 1.4 | 4.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.4 | 52.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 1.4 | 7.1 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 1.4 | 39.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.4 | 7.0 | GO:0046618 | drug export(GO:0046618) |
| 1.4 | 4.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 1.4 | 15.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.4 | 1.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 1.4 | 6.9 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 1.4 | 5.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.4 | 5.4 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 1.3 | 4.0 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 1.3 | 24.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 1.3 | 6.7 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 1.3 | 9.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 1.3 | 6.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.3 | 5.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 1.3 | 11.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 1.3 | 11.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.3 | 20.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 1.3 | 5.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.3 | 11.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 1.2 | 3.7 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.2 | 7.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 1.2 | 3.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.2 | 7.4 | GO:0032899 | regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) |
| 1.2 | 71.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 1.2 | 24.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.2 | 16.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.2 | 14.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 1.2 | 4.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.2 | 16.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 1.2 | 1.2 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 1.1 | 1.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 1.1 | 13.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.1 | 3.4 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 1.1 | 12.3 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 1.1 | 13.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.1 | 3.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.1 | 23.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 1.1 | 19.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 1.1 | 3.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.1 | 6.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.1 | 4.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.1 | 9.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.1 | 9.6 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 1.1 | 6.4 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 1.1 | 7.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.0 | 8.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 1.0 | 8.3 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 1.0 | 9.3 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 1.0 | 15.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 1.0 | 7.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 1.0 | 4.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.0 | 8.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.0 | 4.0 | GO:2000173 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 1.0 | 13.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 1.0 | 19.8 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 1.0 | 29.6 | GO:0097503 | sialylation(GO:0097503) |
| 1.0 | 3.9 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 1.0 | 9.8 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 1.0 | 6.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.0 | 12.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 1.0 | 7.8 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 1.0 | 3.9 | GO:1905165 | regulation of lysosomal protein catabolic process(GO:1905165) |
| 1.0 | 5.8 | GO:0048102 | autophagic cell death(GO:0048102) |
| 1.0 | 6.7 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) midbrain-hindbrain boundary development(GO:0030917) |
| 1.0 | 8.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.0 | 5.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.0 | 2.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.9 | 3.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.9 | 3.7 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.9 | 83.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.9 | 8.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.9 | 2.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.9 | 17.5 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.9 | 4.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.9 | 3.6 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.9 | 19.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.9 | 15.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.9 | 6.3 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.9 | 3.6 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.9 | 7.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.9 | 3.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.9 | 15.7 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.9 | 8.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.9 | 2.6 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.9 | 6.9 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.9 | 5.2 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.9 | 5.2 | GO:2001206 | positive regulation of bone development(GO:1903012) positive regulation of osteoclast development(GO:2001206) |
| 0.9 | 4.3 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.9 | 7.7 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.9 | 8.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.9 | 23.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.8 | 4.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.8 | 7.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.8 | 4.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.8 | 15.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.8 | 2.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.8 | 14.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.8 | 5.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.8 | 32.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.8 | 9.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.8 | 14.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.8 | 8.5 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.8 | 6.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.8 | 4.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.8 | 1.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.8 | 21.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.7 | 4.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 20.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.7 | 5.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.7 | 10.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 20.1 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.7 | 2.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.7 | 6.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.7 | 2.8 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.7 | 6.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.7 | 2.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.7 | 3.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.7 | 5.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.7 | 4.0 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.7 | 4.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.7 | 5.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.7 | 16.4 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.7 | 6.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.7 | 8.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.7 | 7.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.6 | 24.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.6 | 11.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.6 | 17.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.6 | 5.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.6 | 7.5 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.6 | 2.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.6 | 1.8 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.6 | 5.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.6 | 2.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.6 | 1.7 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.6 | 2.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.6 | 2.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.6 | 13.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.6 | 3.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.6 | 0.6 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.6 | 5.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.6 | 1.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.6 | 12.8 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.6 | 28.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.5 | 9.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.5 | 5.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 | 34.2 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.5 | 1.6 | GO:0003011 | diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.5 | 16.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.5 | 3.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.5 | 2.6 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.5 | 6.3 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.5 | 1.6 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.5 | 5.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.5 | 4.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.5 | 5.6 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.5 | 3.1 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.5 | 11.6 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.5 | 12.1 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.5 | 6.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.5 | 9.0 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.5 | 6.0 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.5 | 1.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.5 | 7.4 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.5 | 3.9 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.5 | 11.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.5 | 1.5 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 1.5 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.5 | 3.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.5 | 1.4 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.5 | 5.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 25.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.5 | 27.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.5 | 3.3 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.5 | 2.3 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.5 | 11.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.5 | 1.4 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.5 | 5.9 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.4 | 3.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.4 | 12.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.4 | 7.6 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.4 | 5.3 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.4 | 4.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.4 | 3.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 28.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.4 | 7.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.4 | 10.2 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.4 | 0.8 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.4 | 19.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.4 | 2.1 | GO:0051006 | positive regulation of lipoprotein lipase activity(GO:0051006) |
| 0.4 | 2.0 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.4 | 6.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.4 | 6.5 | GO:0060384 | innervation(GO:0060384) |
| 0.4 | 4.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.4 | 4.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.4 | 0.4 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.4 | 1.6 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.4 | 2.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.4 | 7.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.4 | 5.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.4 | 9.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.4 | 2.7 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.4 | 0.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.4 | 22.9 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.4 | 4.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.4 | 10.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.4 | 1.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.4 | 7.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.4 | 1.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.4 | 1.8 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 1.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 5.2 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.3 | 5.4 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.3 | 3.7 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.3 | 6.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 13.2 | GO:0090102 | cochlea development(GO:0090102) |
| 0.3 | 11.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.3 | 3.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.3 | 2.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 1.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.3 | 17.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.3 | 5.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 9.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 3.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 3.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 3.8 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 2.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 1.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.3 | 5.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 2.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 1.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 | 3.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 2.7 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.3 | 7.6 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 1.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 0.6 | GO:1902022 | lysine transport(GO:0015819) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine transport(GO:1902022) L-lysine import into cell(GO:1903410) |
| 0.3 | 8.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 4.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 1.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 3.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 6.9 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.3 | 4.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.3 | 5.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.3 | 1.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 4.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.3 | GO:0015840 | urea transport(GO:0015840) |
| 0.3 | 1.6 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.3 | 12.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 7.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.3 | 3.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.3 | 7.5 | GO:0009268 | response to pH(GO:0009268) |
| 0.3 | 1.3 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.3 | 5.9 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.3 | 8.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 0.7 | GO:0090500 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.2 | 8.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 4.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 2.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 2.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 4.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 24.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.2 | 0.6 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 | 3.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 4.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.0 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 | 5.2 | GO:0035904 | aorta development(GO:0035904) |
| 0.2 | 0.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 4.1 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.2 | 9.4 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.2 | 4.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 7.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 5.7 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.2 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.2 | 1.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.2 | 0.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 | 6.4 | GO:0008306 | associative learning(GO:0008306) |
| 0.2 | 2.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 18.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 6.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.2 | 0.8 | GO:0003065 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of heart rate by epinephrine(GO:0003065) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.2 | 8.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.2 | 1.9 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 0.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 4.0 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.2 | 1.5 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 3.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.7 | GO:0021675 | nerve development(GO:0021675) |
| 0.1 | 1.4 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 4.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 2.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 1.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 4.0 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.1 | 0.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 3.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 2.0 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 1.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 2.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 3.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 4.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 9.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 6.8 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 6.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 4.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.7 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 2.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 3.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 3.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 1.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.2 | GO:0032417 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 1.7 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 2.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.8 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 2.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 5.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.2 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 1.6 | GO:1902686 | mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.3 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 8.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.8 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.9 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 1.0 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0086004 | regulation of cardiac muscle cell contraction(GO:0086004) |
| 0.0 | 1.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.1 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 1.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 57.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 12.6 | 75.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 12.4 | 62.0 | GO:0031673 | H zone(GO:0031673) |
| 6.4 | 38.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 6.2 | 6.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 4.4 | 13.3 | GO:0008623 | CHRAC(GO:0008623) |
| 4.2 | 25.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 3.8 | 34.4 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 3.8 | 22.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 3.6 | 17.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 3.6 | 14.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 3.6 | 17.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 3.5 | 10.4 | GO:0031251 | PAN complex(GO:0031251) |
| 3.2 | 19.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.9 | 61.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 2.9 | 98.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 2.9 | 17.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.8 | 69.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 2.5 | 38.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 2.3 | 9.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.3 | 22.7 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 2.2 | 6.7 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 2.2 | 8.7 | GO:0070695 | FHF complex(GO:0070695) |
| 2.1 | 6.2 | GO:0032116 | SMC loading complex(GO:0032116) |
| 1.9 | 15.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.8 | 11.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.8 | 5.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.5 | 3.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 1.5 | 7.6 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 1.5 | 36.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 1.4 | 21.7 | GO:0030478 | actin cap(GO:0030478) |
| 1.4 | 8.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.4 | 5.7 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.4 | 9.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.4 | 6.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.4 | 21.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 1.3 | 5.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.3 | 14.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 1.3 | 63.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.2 | 16.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.2 | 27.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.2 | 6.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 1.2 | 6.1 | GO:0044308 | axonal spine(GO:0044308) |
| 1.2 | 15.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 1.1 | 6.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 1.1 | 26.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.1 | 5.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.1 | 7.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.1 | 7.5 | GO:0001939 | female pronucleus(GO:0001939) |
| 1.0 | 9.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.0 | 9.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.0 | 9.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 5.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.0 | 12.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.0 | 5.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.0 | 23.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 1.0 | 109.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 1.0 | 3.9 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 1.0 | 5.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.0 | 11.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 1.0 | 4.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.9 | 3.8 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.9 | 19.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.9 | 29.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.8 | 10.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.8 | 5.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.8 | 34.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.8 | 27.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.8 | 1.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.7 | 14.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 11.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.7 | 7.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.7 | 4.0 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.7 | 7.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 22.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.6 | 3.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.6 | 37.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.6 | 29.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.6 | 6.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.6 | 13.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.6 | 29.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 8.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.6 | 1.7 | GO:0033167 | ARC complex(GO:0033167) |
| 0.6 | 112.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.5 | 8.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.5 | 11.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 10.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 3.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 19.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.5 | 1.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.5 | 9.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 6.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 7.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 2.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.5 | 14.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.5 | 1.9 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.5 | 70.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.5 | 9.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 58.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.5 | 15.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.5 | 13.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 2.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 21.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 9.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.4 | 11.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 21.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 10.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.4 | 6.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 3.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 4.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 13.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.4 | 28.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.4 | 48.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 4.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 1.9 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.4 | 3.6 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 32.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.3 | 2.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 14.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.3 | 5.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 35.0 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 3.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.3 | 49.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 1.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 0.8 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.3 | 3.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 9.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 1.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 2.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 2.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 6.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 5.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 6.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 3.1 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 1.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 7.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 10.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 12.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 34.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 15.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 11.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 1.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 10.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 4.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 4.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 10.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 17.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 29.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 22.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 3.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 45.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.2 | 4.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 28.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 2.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 5.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 9.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 50.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 3.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 163.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 3.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.8 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 3.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 4.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.9 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 116.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 44.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 13.4 | 67.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 10.8 | 32.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 9.2 | 45.8 | GO:0004803 | transposase activity(GO:0004803) |
| 8.8 | 26.4 | GO:0033746 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 6.9 | 27.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 6.8 | 20.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 6.5 | 26.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 6.3 | 25.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 6.0 | 17.9 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 5.8 | 17.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 5.7 | 17.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 5.7 | 17.0 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 5.4 | 21.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 5.2 | 31.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 5.1 | 20.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 4.8 | 67.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 4.8 | 19.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 4.5 | 22.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 4.4 | 17.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 4.0 | 16.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 3.8 | 57.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 3.8 | 11.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 3.7 | 33.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 3.7 | 11.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 3.5 | 10.6 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 3.5 | 14.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 3.4 | 13.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 3.3 | 13.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 3.3 | 16.5 | GO:1903135 | cupric ion binding(GO:1903135) |
| 3.2 | 12.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 3.1 | 18.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 3.1 | 9.3 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 3.1 | 9.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 3.0 | 9.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 3.0 | 21.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 2.9 | 14.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 2.9 | 8.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.9 | 11.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 2.7 | 16.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 2.7 | 32.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.7 | 24.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 2.6 | 34.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 2.6 | 28.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 2.6 | 90.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 2.5 | 19.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 2.4 | 12.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 2.4 | 59.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 2.3 | 13.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.3 | 11.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 2.3 | 9.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 2.3 | 6.8 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 2.2 | 15.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.2 | 6.7 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 2.2 | 32.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 2.2 | 2.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 2.2 | 6.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 2.1 | 18.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 2.0 | 51.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 2.0 | 45.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 2.0 | 9.9 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 2.0 | 90.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 1.9 | 5.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.9 | 13.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 1.9 | 21.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.9 | 19.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.8 | 9.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.8 | 16.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.8 | 7.0 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 1.7 | 10.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.7 | 6.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.7 | 5.2 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 1.7 | 24.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 1.7 | 5.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 1.7 | 5.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.7 | 11.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.7 | 9.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 1.6 | 22.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 1.6 | 6.5 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.6 | 29.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 1.6 | 11.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.6 | 4.8 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol tetrakisphosphate kinase activity(GO:0051765) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 1.6 | 7.9 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 1.6 | 12.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.6 | 9.4 | GO:0070728 | leucine binding(GO:0070728) |
| 1.6 | 14.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.6 | 71.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.6 | 21.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.5 | 20.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 1.5 | 4.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.5 | 7.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.5 | 17.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.5 | 2.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.5 | 5.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.4 | 8.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 1.4 | 21.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.4 | 7.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.4 | 5.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.4 | 4.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 1.4 | 8.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.4 | 6.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.4 | 8.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 1.4 | 40.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.3 | 10.7 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 1.3 | 4.0 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 1.3 | 21.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 1.3 | 31.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 1.3 | 3.9 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 1.3 | 32.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 1.2 | 8.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.2 | 9.7 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 1.2 | 34.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 1.2 | 3.6 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.2 | 9.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 1.2 | 10.5 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 1.2 | 39.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.1 | 31.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.1 | 29.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 1.1 | 3.4 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 1.1 | 3.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.1 | 14.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.1 | 27.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.1 | 14.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 1.1 | 12.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 1.1 | 3.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 1.1 | 16.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 1.1 | 6.3 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 1.0 | 9.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.0 | 19.6 | GO:0031005 | filamin binding(GO:0031005) |
| 1.0 | 6.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.0 | 12.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.0 | 4.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 27.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 1.0 | 7.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.0 | 30.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.0 | 8.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.9 | 2.8 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.9 | 4.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.9 | 3.7 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.9 | 2.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.9 | 19.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.9 | 80.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.9 | 6.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.9 | 21.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.9 | 3.6 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.9 | 2.7 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.9 | 5.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.9 | 2.6 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.9 | 9.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.9 | 4.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.9 | 6.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.8 | 4.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.8 | 4.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.8 | 14.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.8 | 27.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.8 | 5.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.8 | 50.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.8 | 10.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.8 | 4.1 | GO:0015193 | proline:sodium symporter activity(GO:0005298) L-proline transmembrane transporter activity(GO:0015193) |
| 0.8 | 7.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.8 | 3.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.8 | 4.8 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.8 | 26.3 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.8 | 3.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.8 | 24.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.8 | 5.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.8 | 2.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.8 | 5.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.8 | 9.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.8 | 6.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.7 | 15.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 4.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 3.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 5.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.7 | 11.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.7 | 19.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.7 | 10.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.7 | 10.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.7 | 7.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.7 | 4.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.7 | 2.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.7 | 4.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.7 | 3.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.7 | 6.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 8.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.7 | 13.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 15.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 5.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.6 | 1.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.6 | 13.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.6 | 1.9 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.6 | 5.5 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.6 | 6.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.6 | 5.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.6 | 3.5 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.6 | 9.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 4.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.6 | 1.7 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.6 | 4.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 8.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.5 | 8.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 11.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 6.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.5 | 5.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.5 | 6.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 4.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.5 | 1.0 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.5 | 25.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 2.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 4.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 4.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 4.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.5 | 9.0 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.5 | 13.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.5 | 13.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 1.4 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.5 | 1.4 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.5 | 60.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 3.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.5 | 2.3 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.5 | 1.8 | GO:0033265 | choline binding(GO:0033265) |
| 0.5 | 1.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 24.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.4 | 1.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 4.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 1.8 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.4 | 8.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 17.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.4 | 7.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 4.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 3.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 3.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 2.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.4 | 26.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 2.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 6.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 6.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 3.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 9.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 12.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 11.9 | GO:0045502 | dynein binding(GO:0045502) |
| 0.4 | 15.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.4 | 11.8 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.4 | 3.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 4.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.3 | 5.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 2.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.3 | 1.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 3.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 31.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 3.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 14.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 1.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 1.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.3 | 6.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 4.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 5.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 5.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 4.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 6.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 10.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 1.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 11.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.2 | 1.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 24.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.7 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 12.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 16.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 10.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.2 | 4.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 2.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 8.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 2.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 3.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 65.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 6.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 16.6 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.2 | 20.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 0.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 11.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 8.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 0.7 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.2 | 9.9 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.2 | 3.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 7.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 7.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 5.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 5.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 3.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.5 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 6.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.2 | GO:0070325 | lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 16.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.7 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.1 | 1.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 8.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 7.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 2.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 26.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.4 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.1 | 7.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.8 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 2.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.8 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 5.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 88.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.8 | 43.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 1.7 | 34.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 1.6 | 14.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 1.5 | 24.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.4 | 15.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 1.3 | 6.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 1.3 | 18.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 1.2 | 97.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 1.0 | 31.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 1.0 | 43.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.0 | 45.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.9 | 24.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.8 | 23.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.7 | 32.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.7 | 7.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.7 | 59.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.7 | 3.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.6 | 27.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.6 | 20.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.6 | 24.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.6 | 13.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.6 | 23.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.5 | 15.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.5 | 9.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.5 | 136.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 22.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 5.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.4 | 6.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.4 | 3.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 7.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 7.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.4 | 5.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 6.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.4 | 8.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.4 | 6.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 9.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 11.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 10.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 15.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 9.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 4.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 55.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 1.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 8.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 5.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 10.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 20.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 7.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 1.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 10.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 12.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 7.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 7.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 9.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 5.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 19.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 6.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 3.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 1.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 5.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 3.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 54.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 7.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 4.8 | 4.8 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 3.9 | 86.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 2.8 | 35.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 2.7 | 87.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 2.3 | 60.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 2.1 | 6.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 1.9 | 81.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.8 | 10.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.6 | 41.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.6 | 45.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.6 | 34.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 1.5 | 42.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 1.3 | 6.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 1.2 | 8.5 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 1.2 | 16.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.2 | 5.9 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.2 | 26.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 1.2 | 62.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 1.2 | 16.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 1.1 | 19.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 1.0 | 42.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 1.0 | 31.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 1.0 | 22.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.0 | 52.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 1.0 | 18.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.9 | 6.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.9 | 32.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.9 | 15.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.9 | 20.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.8 | 13.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 5.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.7 | 27.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.7 | 2.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.7 | 8.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.7 | 18.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.7 | 11.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.6 | 22.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.6 | 6.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.6 | 19.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.6 | 9.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.6 | 7.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.6 | 20.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 14.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.6 | 18.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 9.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.5 | 6.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.5 | 4.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.5 | 23.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 9.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 19.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.4 | 7.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 10.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 9.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 5.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 4.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.4 | 16.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 68.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.4 | 6.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.4 | 29.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 5.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 6.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 29.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 5.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 10.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 4.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.4 | 9.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.4 | 10.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 5.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 6.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 49.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 21.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 4.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.3 | 7.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.3 | 10.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 9.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.3 | 11.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 34.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.3 | 1.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 17.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 3.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 8.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 4.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 5.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 3.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 5.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 9.2 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.2 | 4.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 15.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 3.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 7.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 15.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 4.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 6.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 4.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 10.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 2.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 14.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 1.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 3.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 6.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 3.1 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 5.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.4 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 2.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 4.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |