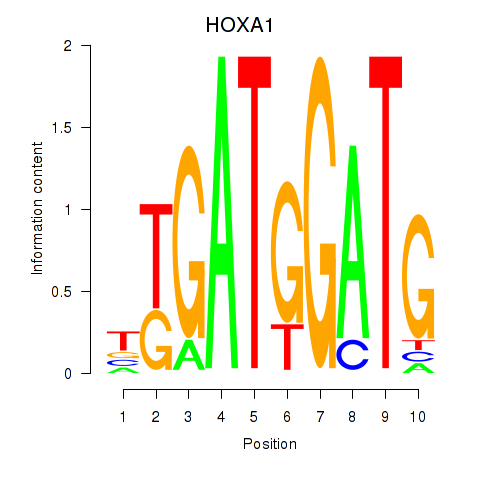
|
chr8_+_103140692
Show fit
|
56.49 |
ENST00000438105.2
ENST00000309982.10
ENST00000297574.6
|
BAALC
|
BAALC binder of MAP3K1 and KLF4
|
|
chr20_-_23637933
Show fit
|
39.95 |
ENST00000398411.5
|
CST3
|
cystatin C
|
|
chr16_+_83968244
Show fit
|
34.18 |
ENST00000305202.9
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr20_-_23637947
Show fit
|
33.11 |
ENST00000376925.8
|
CST3
|
cystatin C
|
|
chr8_-_27611325
Show fit
|
27.65 |
ENST00000523500.5
|
CLU
|
clusterin
|
|
chr5_-_42825884
Show fit
|
25.85 |
ENST00000506577.5
|
SELENOP
|
selenoprotein P
|
|
chr17_-_44911281
Show fit
|
21.83 |
ENST00000638304.1
ENST00000591880.2
ENST00000586125.2
ENST00000639921.1
|
GFAP
|
glial fibrillary acidic protein
|
|
chr8_-_27605271
Show fit
|
20.52 |
ENST00000522098.1
|
CLU
|
clusterin
|
|
chr8_-_27258414
Show fit
|
19.91 |
ENST00000523048.5
|
STMN4
|
stathmin 4
|
|
chr8_-_27611424
Show fit
|
19.68 |
ENST00000405140.7
|
CLU
|
clusterin
|
|
chr20_+_45408276
Show fit
|
18.38 |
ENST00000372710.5
ENST00000443296.1
|
DBNDD2
|
dysbindin domain containing 2
|
|
chr1_+_2073986
Show fit
|
17.87 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta
|
|
chr14_+_99684283
Show fit
|
16.37 |
ENST00000261835.8
|
CYP46A1
|
cytochrome P450 family 46 subfamily A member 1
|
|
chr8_-_27258386
Show fit
|
16.36 |
ENST00000350889.8
ENST00000519997.5
ENST00000519614.5
ENST00000522908.1
ENST00000265770.11
|
STMN4
|
stathmin 4
|
|
chr2_+_24049705
Show fit
|
14.81 |
ENST00000380986.9
ENST00000452109.1
|
FKBP1B
|
FKBP prolyl isomerase 1B
|
|
chrX_-_13817027
Show fit
|
14.73 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr6_+_39792993
Show fit
|
14.29 |
ENST00000538976.5
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr11_-_72642407
Show fit
|
14.21 |
ENST00000376450.7
|
PDE2A
|
phosphodiesterase 2A
|
|
chr6_+_39793008
Show fit
|
13.97 |
ENST00000398904.6
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr11_-_72642450
Show fit
|
13.94 |
ENST00000444035.6
ENST00000544570.5
|
PDE2A
|
phosphodiesterase 2A
|
|
chr4_+_165378998
Show fit
|
13.57 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E
|
|
chr7_+_154305105
Show fit
|
13.40 |
ENST00000332007.7
|
DPP6
|
dipeptidyl peptidase like 6
|
|
chr2_-_50347789
Show fit
|
13.25 |
ENST00000628364.2
|
NRXN1
|
neurexin 1
|
|
chrX_-_73214793
Show fit
|
12.57 |
ENST00000373517.4
|
NAP1L2
|
nucleosome assembly protein 1 like 2
|
|
chr20_+_36154630
Show fit
|
12.38 |
ENST00000338074.7
ENST00000636016.2
ENST00000373945.5
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr4_+_113049616
Show fit
|
12.20 |
ENST00000504454.5
ENST00000357077.9
ENST00000394537.7
ENST00000672779.1
ENST00000264366.10
|
ANK2
|
ankyrin 2
|
|
chr7_-_137343688
Show fit
|
11.62 |
ENST00000348225.7
|
PTN
|
pleiotrophin
|
|
chr17_-_1229706
Show fit
|
11.42 |
ENST00000574139.7
|
ABR
|
ABR activator of RhoGEF and GTPase
|
|
chr16_+_15502266
Show fit
|
11.27 |
ENST00000452191.6
|
BMERB1
|
bMERB domain containing 1
|
|
chr11_-_5227063
Show fit
|
11.09 |
ENST00000335295.4
ENST00000485743.1
ENST00000647020.1
|
HBB
|
hemoglobin subunit beta
|
|
chr3_+_159839847
Show fit
|
10.52 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_+_24049673
Show fit
|
10.31 |
ENST00000380991.8
|
FKBP1B
|
FKBP prolyl isomerase 1B
|
|
chr3_+_150546765
Show fit
|
10.18 |
ENST00000406576.7
ENST00000460851.6
ENST00000482093.5
ENST00000273435.9
|
EIF2A
|
eukaryotic translation initiation factor 2A
|
|
chr16_+_23835946
Show fit
|
9.99 |
ENST00000321728.12
ENST00000643927.1
|
PRKCB
|
protein kinase C beta
|
|
chr2_+_230864921
Show fit
|
9.87 |
ENST00000326427.11
ENST00000335005.10
ENST00000326407.10
|
ITM2C
|
integral membrane protein 2C
|
|
chr7_-_137343752
Show fit
|
9.85 |
ENST00000393083.2
|
PTN
|
pleiotrophin
|
|
chr4_+_113049479
Show fit
|
9.84 |
ENST00000671727.1
ENST00000671762.1
ENST00000672366.1
ENST00000672502.1
ENST00000672045.1
ENST00000672251.1
ENST00000672854.1
|
ANK2
|
ankyrin 2
|
|
chr4_+_153257339
Show fit
|
9.68 |
ENST00000676374.1
ENST00000676196.1
ENST00000674935.1
ENST00000674769.1
ENST00000674896.1
ENST00000676191.1
ENST00000675312.1
ENST00000675456.1
ENST00000674847.1
ENST00000675977.1
ENST00000676264.1
ENST00000674726.1
ENST00000676252.1
ENST00000674730.1
ENST00000675738.1
ENST00000482578.3
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_+_26188836
Show fit
|
9.18 |
ENST00000672621.1
|
ANO3
|
anoctamin 3
|
|
chr4_-_86101922
Show fit
|
8.79 |
ENST00000472236.5
ENST00000641881.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr11_-_104164126
Show fit
|
8.52 |
ENST00000393158.7
|
PDGFD
|
platelet derived growth factor D
|
|
chr1_+_65264694
Show fit
|
8.34 |
ENST00000263441.11
ENST00000395325.7
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6
|
|
chr3_+_159852933
Show fit
|
8.27 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_-_44361555
Show fit
|
7.76 |
ENST00000409957.5
|
PREPL
|
prolyl endopeptidase like
|
|
chr8_-_90082871
Show fit
|
7.75 |
ENST00000265431.7
|
CALB1
|
calbindin 1
|
|
chr2_-_50347710
Show fit
|
7.54 |
ENST00000342183.9
ENST00000401710.5
|
NRXN1
|
neurexin 1
|
|
chr4_+_70721953
Show fit
|
7.52 |
ENST00000381006.8
ENST00000226328.8
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr12_-_91146195
Show fit
|
7.30 |
ENST00000548218.1
|
DCN
|
decorin
|
|
chr5_+_140834230
Show fit
|
7.16 |
ENST00000356878.5
ENST00000525929.2
|
PCDHA7
|
protocadherin alpha 7
|
|
chr6_+_32024278
Show fit
|
6.73 |
ENST00000647698.1
|
C4B
|
complement C4B (Chido blood group)
|
|
chr20_+_9514562
Show fit
|
6.34 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5
|
|
chr6_+_17393576
Show fit
|
6.18 |
ENST00000229922.7
ENST00000611958.4
|
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2
|
|
chr11_-_104164361
Show fit
|
5.98 |
ENST00000302251.9
|
PDGFD
|
platelet derived growth factor D
|
|
chr6_+_17393505
Show fit
|
5.94 |
ENST00000616440.4
|
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2
|
|
chr1_+_205504592
Show fit
|
5.89 |
ENST00000506784.5
ENST00000360066.6
|
CDK18
|
cyclin dependent kinase 18
|
|
chr6_+_17393607
Show fit
|
5.79 |
ENST00000489374.5
ENST00000378990.6
|
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2
|
|
chr10_+_7703340
Show fit
|
5.54 |
ENST00000429820.5
ENST00000379587.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chrM_+_4467
Show fit
|
5.52 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2
|
|
chr1_-_13201409
Show fit
|
5.50 |
ENST00000625019.3
|
PRAMEF13
|
PRAME family member 13
|
|
chr11_+_125904467
Show fit
|
5.50 |
ENST00000263576.11
ENST00000530414.5
ENST00000530129.6
|
DDX25
|
DEAD-box helicase 25
|
|
chr12_+_12725897
Show fit
|
5.24 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr9_-_94640130
Show fit
|
5.09 |
ENST00000414122.1
|
FBP1
|
fructose-bisphosphatase 1
|
|
chrX_-_57910458
Show fit
|
4.88 |
ENST00000358697.6
|
ZXDA
|
zinc finger X-linked duplicated A
|
|
chr9_-_94640248
Show fit
|
4.88 |
ENST00000415431.5
|
FBP1
|
fructose-bisphosphatase 1
|
|
chr5_-_784691
Show fit
|
4.85 |
ENST00000508859.8
ENST00000652055.1
ENST00000651083.1
|
ZDHHC11B
|
zinc finger DHHC-type containing 11B
|
|
chr3_+_38496467
Show fit
|
4.83 |
ENST00000453767.1
|
EXOG
|
exo/endonuclease G
|
|
chr1_-_13347134
Show fit
|
4.79 |
ENST00000334600.7
|
PRAMEF14
|
PRAME family member 14
|
|
chr7_-_22193824
Show fit
|
4.73 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5
|
|
chr8_+_22275309
Show fit
|
4.72 |
ENST00000356766.11
ENST00000521356.5
|
PIWIL2
|
piwi like RNA-mediated gene silencing 2
|
|
chr9_-_10612966
Show fit
|
4.72 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D
|
|
chr6_-_13487593
Show fit
|
4.62 |
ENST00000379287.4
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr2_-_152098810
Show fit
|
4.54 |
ENST00000636442.1
ENST00000638005.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4
|
|
chr9_-_83956677
Show fit
|
4.52 |
ENST00000376344.8
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr7_-_22193728
Show fit
|
4.39 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5
|
|
chr8_+_142449430
Show fit
|
4.36 |
ENST00000643448.1
ENST00000517894.5
|
ADGRB1
|
adhesion G protein-coupled receptor B1
|
|
chr7_+_80135694
Show fit
|
4.36 |
ENST00000457358.7
|
GNAI1
|
G protein subunit alpha i1
|
|
chr3_-_129893551
Show fit
|
4.31 |
ENST00000505616.5
ENST00000426664.6
ENST00000648771.1
ENST00000393238.8
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chrX_-_6227180
Show fit
|
4.31 |
ENST00000381093.6
|
NLGN4X
|
neuroligin 4 X-linked
|
|
chr1_-_201399525
Show fit
|
4.16 |
ENST00000367313.4
|
LAD1
|
ladinin 1
|
|
chr3_+_4493442
Show fit
|
4.13 |
ENST00000456211.8
ENST00000443694.5
ENST00000648266.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr10_+_7703300
Show fit
|
4.13 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr19_+_840991
Show fit
|
4.12 |
ENST00000234347.10
|
PRTN3
|
proteinase 3
|
|
chr3_+_4493471
Show fit
|
4.11 |
ENST00000544951.6
ENST00000650294.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr6_+_151239951
Show fit
|
3.95 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12
|
|
chr16_+_75222609
Show fit
|
3.94 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1
|
|
chr11_-_89065969
Show fit
|
3.92 |
ENST00000305447.5
|
GRM5
|
glutamate metabotropic receptor 5
|
|
chr3_+_4493340
Show fit
|
3.88 |
ENST00000357086.10
ENST00000354582.12
ENST00000649015.2
ENST00000467056.6
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr5_-_20575850
Show fit
|
3.85 |
ENST00000507958.5
|
CDH18
|
cadherin 18
|
|
chr19_+_13764502
Show fit
|
3.82 |
ENST00000040663.8
ENST00000319545.12
|
MRI1
|
methylthioribose-1-phosphate isomerase 1
|
|
chrX_+_57592011
Show fit
|
3.80 |
ENST00000374888.3
|
ZXDB
|
zinc finger X-linked duplicated B
|
|
chr12_-_54984667
Show fit
|
3.79 |
ENST00000524668.5
ENST00000533607.1
ENST00000449076.6
|
TESPA1
|
thymocyte expressed, positive selection associated 1
|
|
chr5_+_134526100
Show fit
|
3.65 |
ENST00000395003.5
|
JADE2
|
jade family PHD finger 2
|
|
chr10_+_94089034
Show fit
|
3.60 |
ENST00000676102.1
ENST00000371385.8
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr13_+_35476740
Show fit
|
3.55 |
ENST00000537702.5
|
NBEA
|
neurobeachin
|
|
chr1_-_7853054
Show fit
|
3.51 |
ENST00000361696.10
|
UTS2
|
urotensin 2
|
|
chr5_-_137736066
Show fit
|
3.47 |
ENST00000309755.9
|
KLHL3
|
kelch like family member 3
|
|
chr9_+_33795551
Show fit
|
3.46 |
ENST00000379405.4
|
PRSS3
|
serine protease 3
|
|
chr19_+_5681000
Show fit
|
3.37 |
ENST00000581893.5
ENST00000411793.6
ENST00000301382.8
ENST00000581773.5
ENST00000339423.7
ENST00000423665.6
ENST00000583928.5
ENST00000342970.6
ENST00000422535.6
ENST00000581521.5
|
HSD11B1L
|
hydroxysteroid 11-beta dehydrogenase 1 like
|
|
chr5_-_126595185
Show fit
|
3.37 |
ENST00000510111.6
ENST00000635851.1
ENST00000637964.1
ENST00000413020.6
ENST00000637782.1
ENST00000409134.8
ENST00000637272.1
ENST00000636879.1
ENST00000636743.1
ENST00000636886.1
ENST00000509270.2
|
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1
|
|
chr6_+_101393699
Show fit
|
3.35 |
ENST00000369134.9
ENST00000684068.1
ENST00000683903.1
ENST00000681975.1
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2
|
|
chr17_-_15598797
Show fit
|
3.34 |
ENST00000354433.7
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr16_-_66550112
Show fit
|
3.25 |
ENST00000544898.6
ENST00000620035.5
ENST00000545043.6
|
TK2
|
thymidine kinase 2
|
|
chr7_+_94509793
Show fit
|
3.06 |
ENST00000297273.9
|
CASD1
|
CAS1 domain containing 1
|
|
chr5_-_134367144
Show fit
|
3.05 |
ENST00000265334.9
|
CDKL3
|
cyclin dependent kinase like 3
|
|
chr17_-_75405486
Show fit
|
2.98 |
ENST00000392562.5
|
GRB2
|
growth factor receptor bound protein 2
|
|
chr12_-_101210232
Show fit
|
2.98 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8
|
|
chr19_-_38878247
Show fit
|
2.93 |
ENST00000591812.2
|
RINL
|
Ras and Rab interactor like
|
|
chr16_-_10942443
Show fit
|
2.91 |
ENST00000570440.2
ENST00000331808.5
|
DEXI
|
Dexi homolog
|
|
chrX_-_43973382
Show fit
|
2.77 |
ENST00000642620.1
ENST00000647044.1
|
NDP
|
norrin cystine knot growth factor NDP
|
|
chrX_+_71223216
Show fit
|
2.67 |
ENST00000361726.7
|
GJB1
|
gap junction protein beta 1
|
|
chr11_-_70717994
Show fit
|
2.65 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr17_-_15598618
Show fit
|
2.64 |
ENST00000583965.5
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr12_-_84912783
Show fit
|
2.63 |
ENST00000680892.1
ENST00000266682.10
ENST00000680714.1
ENST00000552192.5
|
SLC6A15
|
solute carrier family 6 member 15
|
|
chr16_+_29662923
Show fit
|
2.63 |
ENST00000395389.2
|
SPN
|
sialophorin
|
|
chr12_+_124993633
Show fit
|
2.57 |
ENST00000341446.9
ENST00000671775.2
|
BRI3BP
|
BRI3 binding protein
|
|
chr12_+_8123899
Show fit
|
2.56 |
ENST00000641376.1
|
CLEC4A
|
C-type lectin domain family 4 member A
|
|
chr17_-_3292600
Show fit
|
2.51 |
ENST00000615105.1
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1
|
|
chr17_-_27893339
Show fit
|
2.48 |
ENST00000460380.6
ENST00000379102.8
ENST00000508862.5
ENST00000582441.1
|
LYRM9
ENSG00000266202.1
|
LYR motif containing 9
novel protein
|
|
chr1_-_110519175
Show fit
|
2.38 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10
|
|
chr9_-_6645712
Show fit
|
2.37 |
ENST00000321612.8
|
GLDC
|
glycine decarboxylase
|
|
chr1_-_100249815
Show fit
|
2.35 |
ENST00000370131.3
ENST00000681617.1
ENST00000681780.1
ENST00000370132.8
|
DBT
|
dihydrolipoamide branched chain transacylase E2
|
|
chr13_+_91398613
Show fit
|
2.35 |
ENST00000377067.9
|
GPC5
|
glypican 5
|
|
chr6_+_151240368
Show fit
|
2.35 |
ENST00000253332.5
|
AKAP12
|
A-kinase anchoring protein 12
|
|
chrX_+_15790446
Show fit
|
2.31 |
ENST00000380308.7
ENST00000307771.8
|
ZRSR2
|
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2
|
|
chr12_+_57216779
Show fit
|
2.26 |
ENST00000349394.6
|
NXPH4
|
neurexophilin 4
|
|
chr16_-_67393486
Show fit
|
2.22 |
ENST00000562206.1
ENST00000393957.7
ENST00000290942.9
|
TPPP3
|
tubulin polymerization promoting protein family member 3
|
|
chrX_+_54808359
Show fit
|
2.18 |
ENST00000375058.5
ENST00000375060.5
|
MAGED2
|
MAGE family member D2
|
|
chr16_+_29663219
Show fit
|
2.13 |
ENST00000436527.5
ENST00000360121.4
ENST00000652691.1
ENST00000449759.2
|
SPN
QPRT
|
sialophorin
quinolinate phosphoribosyltransferase
|
|
chr2_-_60550900
Show fit
|
2.12 |
ENST00000643222.1
ENST00000643459.1
ENST00000489516.7
|
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A
|
|
chr8_-_40897814
Show fit
|
2.12 |
ENST00000297737.11
ENST00000315769.11
|
ZMAT4
|
zinc finger matrin-type 4
|
|
chr1_+_162632454
Show fit
|
2.11 |
ENST00000367921.8
ENST00000367922.7
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr12_-_84912705
Show fit
|
2.10 |
ENST00000679933.1
ENST00000680260.1
ENST00000551010.2
ENST00000679453.1
ENST00000681281.1
|
SLC6A15
|
solute carrier family 6 member 15
|
|
chr5_-_126595237
Show fit
|
2.09 |
ENST00000637206.1
ENST00000553117.5
|
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1
|
|
chr6_+_13013513
Show fit
|
2.07 |
ENST00000675203.1
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr22_+_35299800
Show fit
|
2.07 |
ENST00000456128.5
ENST00000411850.5
ENST00000425375.5
ENST00000449058.7
|
TOM1
|
target of myb1 membrane trafficking protein
|
|
chr17_+_70075215
Show fit
|
2.04 |
ENST00000283936.5
ENST00000615244.4
ENST00000392671.6
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16
|
|
chr1_+_156154371
Show fit
|
2.04 |
ENST00000368282.1
|
SEMA4A
|
semaphorin 4A
|
|
chr19_-_40226682
Show fit
|
2.03 |
ENST00000430325.7
ENST00000599263.6
|
CCNP
|
cyclin P
|
|
chr1_-_100895132
Show fit
|
2.02 |
ENST00000535414.5
|
EXTL2
|
exostosin like glycosyltransferase 2
|
|
chr13_-_77919390
Show fit
|
1.97 |
ENST00000475537.2
ENST00000646605.1
|
EDNRB
|
endothelin receptor type B
|
|
chr6_-_47042260
Show fit
|
1.81 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1
|
|
chr1_+_3698027
Show fit
|
1.80 |
ENST00000378290.4
|
TP73
|
tumor protein p73
|
|
chr19_+_15049469
Show fit
|
1.79 |
ENST00000427043.4
|
CASP14
|
caspase 14
|
|
chr22_-_38794111
Show fit
|
1.78 |
ENST00000406622.5
ENST00000216068.9
ENST00000406199.3
|
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2
dynein axonemal light chain 4
|
|
chr3_+_44874606
Show fit
|
1.76 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4
|
|
chr17_+_28473278
Show fit
|
1.73 |
ENST00000444914.7
|
SLC13A2
|
solute carrier family 13 member 2
|
|
chrX_+_37780049
Show fit
|
1.70 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain
|
|
chr12_-_21774688
Show fit
|
1.70 |
ENST00000240662.3
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8
|
|
chr11_-_89063631
Show fit
|
1.68 |
ENST00000455756.6
|
GRM5
|
glutamate metabotropic receptor 5
|
|
chr1_-_112956063
Show fit
|
1.68 |
ENST00000538576.5
ENST00000369626.8
ENST00000458229.6
|
SLC16A1
|
solute carrier family 16 member 1
|
|
chr15_-_58014097
Show fit
|
1.63 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2
|
|
chr11_+_68460712
Show fit
|
1.62 |
ENST00000528635.5
ENST00000533127.5
ENST00000529907.5
ENST00000529344.5
ENST00000534534.5
ENST00000524845.5
ENST00000393800.7
ENST00000265637.8
ENST00000524904.5
ENST00000393801.7
ENST00000265636.9
ENST00000529710.5
|
PPP6R3
|
protein phosphatase 6 regulatory subunit 3
|
|
chr7_-_117323041
Show fit
|
1.57 |
ENST00000491214.1
ENST00000265441.8
|
WNT2
|
Wnt family member 2
|
|
chr22_-_50261543
Show fit
|
1.55 |
ENST00000395778.3
ENST00000215659.13
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chrX_+_83508284
Show fit
|
1.53 |
ENST00000644024.2
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr1_-_3796478
Show fit
|
1.43 |
ENST00000378251.3
|
LRRC47
|
leucine rich repeat containing 47
|
|
chr9_-_21368057
Show fit
|
1.41 |
ENST00000449498.2
|
IFNA13
|
interferon alpha 13
|
|
chr11_-_2140967
Show fit
|
1.38 |
ENST00000381389.5
|
IGF2
|
insulin like growth factor 2
|
|
chr2_-_199458689
Show fit
|
1.37 |
ENST00000443023.5
|
SATB2
|
SATB homeobox 2
|
|
chr20_-_63537361
Show fit
|
1.33 |
ENST00000217185.3
ENST00000542869.3
|
PTK6
|
protein tyrosine kinase 6
|
|
chr21_+_31873010
Show fit
|
1.32 |
ENST00000270112.7
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr10_+_18400562
Show fit
|
1.32 |
ENST00000377315.5
ENST00000650685.1
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr5_-_134367070
Show fit
|
1.31 |
ENST00000521118.5
|
CDKL3
|
cyclin dependent kinase like 3
|
|
chr17_-_3595831
Show fit
|
1.27 |
ENST00000399759.7
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1
|
|
chr1_+_17308194
Show fit
|
1.21 |
ENST00000375453.5
ENST00000375448.4
|
PADI4
|
peptidyl arginine deiminase 4
|
|
chr7_-_101165558
Show fit
|
1.19 |
ENST00000611537.1
ENST00000249330.3
|
VGF
|
VGF nerve growth factor inducible
|
|
chr14_+_24232921
Show fit
|
1.17 |
ENST00000557854.5
ENST00000399440.7
ENST00000559104.5
ENST00000456667.7
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr7_-_44141285
Show fit
|
1.16 |
ENST00000458240.5
ENST00000223364.7
|
MYL7
|
myosin light chain 7
|
|
chr11_+_3645105
Show fit
|
1.14 |
ENST00000250693.2
|
ART1
|
ADP-ribosyltransferase 1
|
|
chr7_-_44141074
Show fit
|
1.13 |
ENST00000457314.5
ENST00000447951.1
ENST00000431007.1
|
MYL7
|
myosin light chain 7
|
|
chr9_-_5304713
Show fit
|
1.10 |
ENST00000381627.4
|
RLN2
|
relaxin 2
|
|
chr1_-_23217482
Show fit
|
1.09 |
ENST00000374619.2
|
HTR1D
|
5-hydroxytryptamine receptor 1D
|
|
chr1_+_18630839
Show fit
|
1.03 |
ENST00000420770.7
|
PAX7
|
paired box 7
|
|
chr14_+_24310134
Show fit
|
1.02 |
ENST00000533293.2
ENST00000543919.1
|
LTB4R2
|
leukotriene B4 receptor 2
|
|
chrX_+_15507302
Show fit
|
1.01 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr9_-_113221243
Show fit
|
1.01 |
ENST00000238256.8
|
FKBP15
|
FKBP prolyl isomerase family member 15
|
|
chrX_+_54808334
Show fit
|
1.00 |
ENST00000218439.8
|
MAGED2
|
MAGE family member D2
|
|
chr16_+_71626175
Show fit
|
0.99 |
ENST00000268485.8
ENST00000565261.1
ENST00000299952.4
|
MARVELD3
|
MARVEL domain containing 3
|
|
chrY_-_13986473
Show fit
|
0.97 |
ENST00000250825.5
|
VCY
|
variable charge Y-linked
|
|
chr16_+_30069539
Show fit
|
0.96 |
ENST00000565355.1
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chr17_-_44199834
Show fit
|
0.93 |
ENST00000587097.6
|
ATXN7L3
|
ataxin 7 like 3
|
|
chr1_+_36156096
Show fit
|
0.93 |
ENST00000474796.2
ENST00000373150.8
ENST00000373151.6
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr11_-_101583503
Show fit
|
0.92 |
ENST00000348423.8
ENST00000360497.4
ENST00000532133.5
|
TRPC6
|
transient receptor potential cation channel subfamily C member 6
|
|
chrX_+_48597482
Show fit
|
0.92 |
ENST00000218056.9
ENST00000376729.10
|
WDR13
|
WD repeat domain 13
|
|
chr10_-_103855406
Show fit
|
0.89 |
ENST00000355946.6
ENST00000369774.8
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr16_+_16379055
Show fit
|
0.87 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family member A7
|
|
chr13_+_29428603
Show fit
|
0.87 |
ENST00000380808.6
|
MTUS2
|
microtubule associated scaffold protein 2
|
|
chr13_-_20161038
Show fit
|
0.84 |
ENST00000241125.4
|
GJA3
|
gap junction protein alpha 3
|
|
chr19_-_42423100
Show fit
|
0.84 |
ENST00000597001.1
|
LIPE
|
lipase E, hormone sensitive type
|
|
chr12_-_96400365
Show fit
|
0.83 |
ENST00000261211.8
ENST00000543119.6
|
CDK17
|
cyclin dependent kinase 17
|
|
chr1_-_24143112
Show fit
|
0.82 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
|
chr16_-_28471175
Show fit
|
0.79 |
ENST00000435324.3
|
NPIPB7
|
nuclear pore complex interacting protein family member B7
|
|
chr17_-_46818680
Show fit
|
0.74 |
ENST00000225512.6
|
WNT3
|
Wnt family member 3
|
|
chr7_+_139829242
Show fit
|
0.70 |
ENST00000455353.6
ENST00000458722.6
ENST00000448866.7
ENST00000411653.6
|
TBXAS1
|
thromboxane A synthase 1
|
|
chr19_+_47019800
Show fit
|
0.69 |
ENST00000602212.6
ENST00000602189.5
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr19_-_49451793
Show fit
|
0.69 |
ENST00000262265.10
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr1_+_43300971
Show fit
|
0.69 |
ENST00000372476.8
ENST00000538015.1
|
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1
|
|
chr16_-_55833186
Show fit
|
0.68 |
ENST00000361503.8
ENST00000422046.6
|
CES1
|
carboxylesterase 1
|
|
chr8_-_96235533
Show fit
|
0.68 |
ENST00000518406.5
ENST00000287022.10
ENST00000523920.1
|
UQCRB
|
ubiquinol-cytochrome c reductase binding protein
|
|
chr6_+_25754699
Show fit
|
0.65 |
ENST00000439485.6
ENST00000377905.9
|
SLC17A4
|
solute carrier family 17 member 4
|